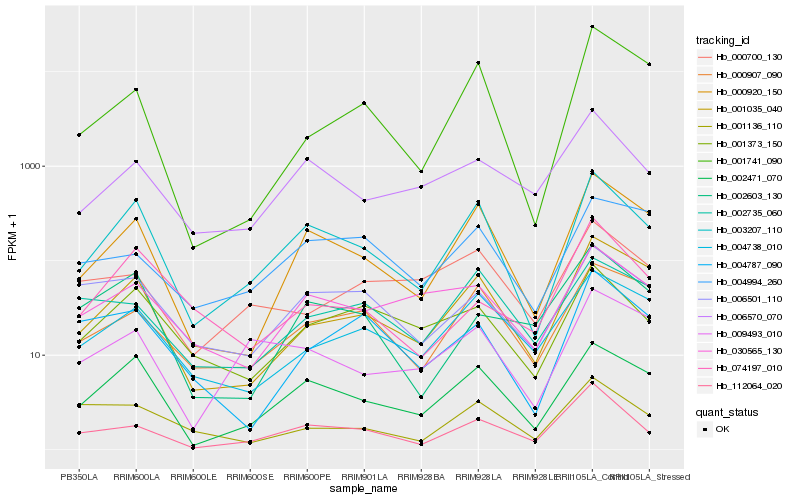
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112064_020 |
0.0 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 2 |
Hb_006570_070 |
0.1647977346 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 3 |
Hb_003207_110 |
0.1680721817 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 4 |
Hb_000920_150 |
0.1824860165 |
- |
- |
annexin [Manihot esculenta] |
| 5 |
Hb_006501_110 |
0.19227635 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002603_130 |
0.1980411707 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 7 |
Hb_000907_090 |
0.2075434486 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001035_040 |
0.2114141751 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_002471_070 |
0.2116172391 |
- |
- |
Tartrate-resistant acid phosphatase type 5 precursor, putative [Ricinus communis] |
| 10 |
Hb_004787_090 |
0.2116842984 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 11 |
Hb_000700_130 |
0.2127676519 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001373_150 |
0.2128645801 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 13 |
Hb_001136_110 |
0.2142183206 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38) isoform X1 [Jatropha curcas] |
| 14 |
Hb_004738_010 |
0.2164812091 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 15 |
Hb_002735_060 |
0.2171988839 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 16 |
Hb_030565_130 |
0.2186553554 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 17 |
Hb_009493_010 |
0.2287757032 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 18 |
Hb_074197_010 |
0.2294402883 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 19 |
Hb_001741_090 |
0.2298157131 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 20 |
Hb_004994_260 |
0.229901336 |
- |
- |
Remorin, putative [Ricinus communis] |