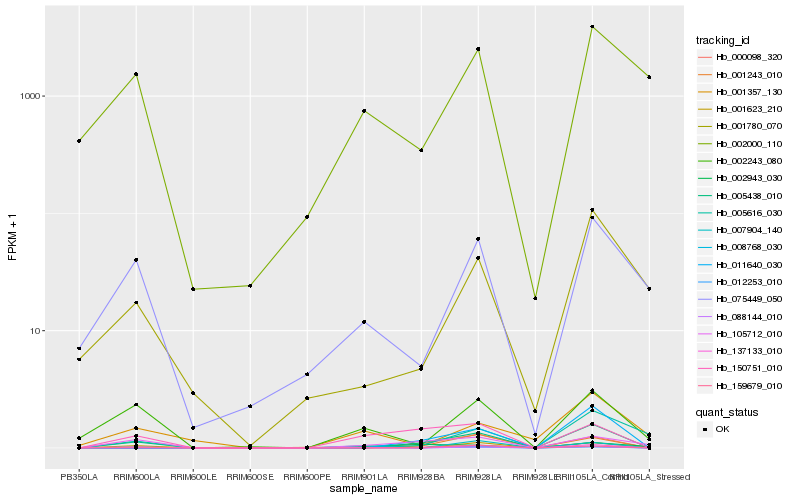
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012253_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_005438_010 |
0.2025034075 |
- |
- |
PREDICTED: uncharacterized protein LOC104902779 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_150751_010 |
0.2669940047 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 4 |
Hb_011640_030 |
0.2881479641 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 5 |
Hb_159679_010 |
0.2906717133 |
- |
- |
PREDICTED: uncharacterized protein LOC105786838 [Gossypium raimondii] |
| 6 |
Hb_001623_210 |
0.2953219356 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_008768_030 |
0.2971114784 |
- |
- |
- |
| 8 |
Hb_002243_080 |
0.2971977016 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 9 |
Hb_075449_050 |
0.3153711859 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 10 |
Hb_105712_010 |
0.3159915178 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 11 |
Hb_001780_070 |
0.3233576188 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 12 |
Hb_001357_130 |
0.3291871763 |
- |
- |
hypothetical protein JCGZ_00933 [Jatropha curcas] |
| 13 |
Hb_002943_030 |
0.329869072 |
- |
- |
- |
| 14 |
Hb_007904_140 |
0.3327307208 |
- |
- |
- |
| 15 |
Hb_002000_110 |
0.3372922665 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 16 |
Hb_001243_010 |
0.3396759261 |
- |
- |
PREDICTED: dirigent protein 4-like [Populus euphratica] |
| 17 |
Hb_000098_320 |
0.3400920059 |
- |
- |
- |
| 18 |
Hb_137133_010 |
0.3402850003 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 19 |
Hb_005616_030 |
0.3409159134 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 20 |
Hb_088144_010 |
0.341103108 |
- |
- |
cellulose synthase, putative [Ricinus communis] |