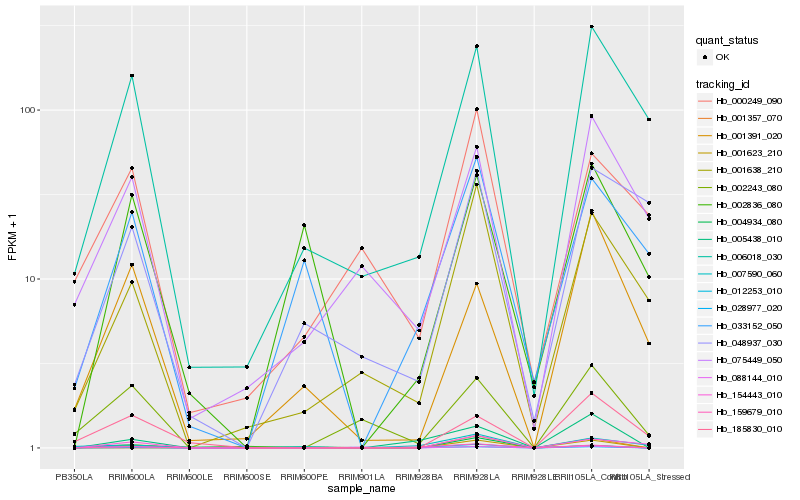
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_210 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 2 |
Hb_001638_210 |
0.2689966412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006018_030 |
0.2819335428 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 4 |
Hb_159679_010 |
0.2883648192 |
- |
- |
PREDICTED: uncharacterized protein LOC105786838 [Gossypium raimondii] |
| 5 |
Hb_002836_080 |
0.2907077762 |
- |
- |
- |
| 6 |
Hb_012253_010 |
0.2953219356 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 7 |
Hb_005438_010 |
0.2962513837 |
- |
- |
PREDICTED: uncharacterized protein LOC104902779 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_001391_020 |
0.2991228092 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 9 |
Hb_002243_080 |
0.2998062675 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 10 |
Hb_154443_010 |
0.3033866595 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 11 |
Hb_075449_050 |
0.3047546421 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 12 |
Hb_033152_050 |
0.3055699455 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 13 |
Hb_000249_090 |
0.327023328 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 14 |
Hb_028977_020 |
0.3329130022 |
- |
- |
hypothetical protein JCGZ_21319 [Jatropha curcas] |
| 15 |
Hb_088144_010 |
0.3333657109 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 16 |
Hb_007590_060 |
0.3347040547 |
- |
- |
- |
| 17 |
Hb_048937_030 |
0.3427318559 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 18 |
Hb_185830_010 |
0.343881886 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1 [Jatropha curcas] |
| 19 |
Hb_004934_080 |
0.3461289509 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 20 |
Hb_001357_070 |
0.3465022427 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |