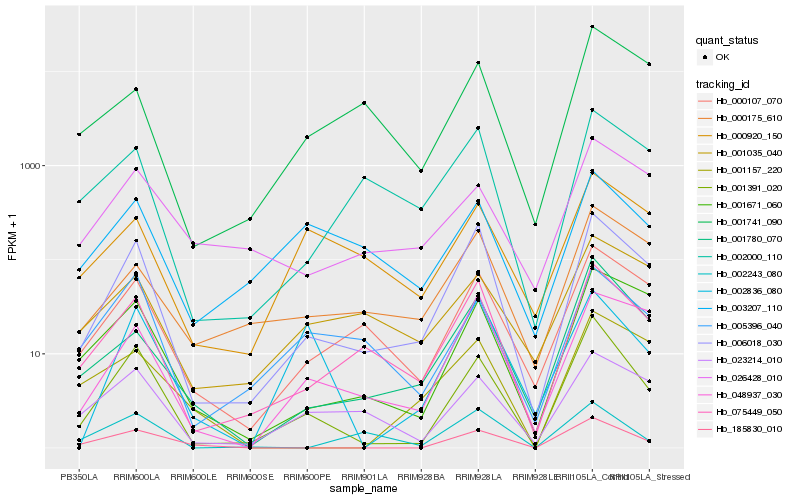
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001391_020 |
0.0 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 2 |
Hb_006018_030 |
0.1689760691 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 3 |
Hb_185830_010 |
0.1826926995 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1 [Jatropha curcas] |
| 4 |
Hb_075449_050 |
0.2034328584 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 5 |
Hb_001671_060 |
0.2176610455 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 6 |
Hb_001780_070 |
0.2208408558 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 7 |
Hb_000107_070 |
0.2257956831 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 8 |
Hb_005396_040 |
0.2277942544 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_023214_010 |
0.2293979427 |
- |
- |
aberrant lateral root formation 1 family protein [Populus trichocarpa] |
| 10 |
Hb_026428_010 |
0.2547891508 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 11 |
Hb_003207_110 |
0.2564060953 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 12 |
Hb_002836_080 |
0.258682209 |
- |
- |
- |
| 13 |
Hb_000175_610 |
0.2634363024 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 14 |
Hb_048937_030 |
0.264138146 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 15 |
Hb_000920_150 |
0.2662185034 |
- |
- |
annexin [Manihot esculenta] |
| 16 |
Hb_002243_080 |
0.2709454362 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 17 |
Hb_001157_220 |
0.2718928568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001741_090 |
0.2721393368 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 19 |
Hb_002000_110 |
0.2721639895 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 20 |
Hb_001035_040 |
0.2739293867 |
- |
- |
unnamed protein product [Coffea canephora] |