| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
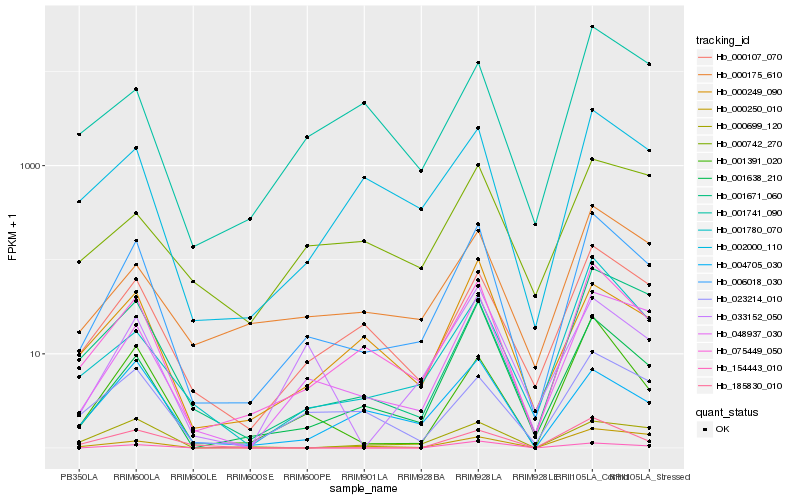
Hb_006018_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 2 |
Hb_075449_050 |
0.1110308376 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 3 |
Hb_048937_030 |
0.136694683 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 4 |
Hb_000107_070 |
0.1567558062 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 5 |
Hb_001391_020 |
0.1689760691 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 6 |
Hb_001638_210 |
0.1724796259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001671_060 |
0.173626106 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 8 |
Hb_002000_110 |
0.1754756084 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 9 |
Hb_000175_610 |
0.184799132 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001780_070 |
0.192071129 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 11 |
Hb_033152_050 |
0.1958774678 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 12 |
Hb_023214_010 |
0.1963473027 |
- |
- |
aberrant lateral root formation 1 family protein [Populus trichocarpa] |
| 13 |
Hb_000249_090 |
0.1988957019 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 14 |
Hb_004705_030 |
0.2011647404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000699_120 |
0.202088955 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 16 |
Hb_154443_010 |
0.2062923698 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 17 |
Hb_185830_010 |
0.2138555438 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1 [Jatropha curcas] |
| 18 |
Hb_000742_270 |
0.2203354169 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 19 |
Hb_001741_090 |
0.2205370142 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 20 |
Hb_000250_010 |
0.2210292591 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |