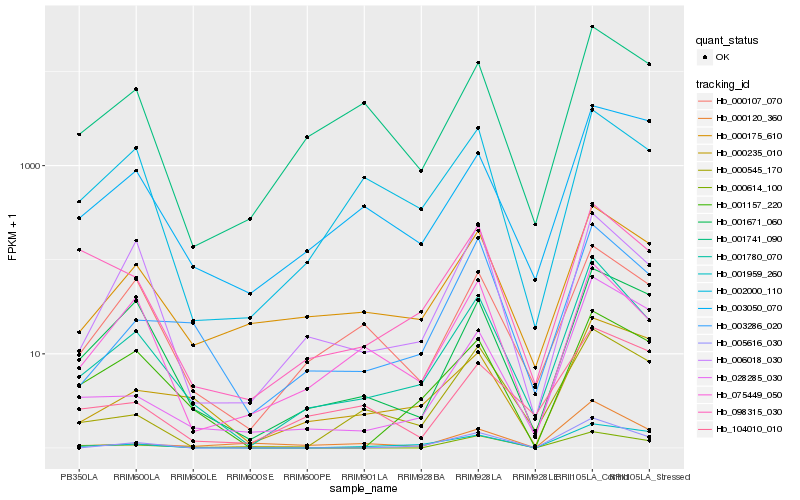
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001780_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 2 |
Hb_000175_610 |
0.1637785334 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 3 |
Hb_003286_020 |
0.1680808215 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 4 |
Hb_005616_030 |
0.1806469239 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 5 |
Hb_001741_090 |
0.1827342176 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 6 |
Hb_075449_050 |
0.185361554 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 7 |
Hb_000107_070 |
0.1859875291 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 8 |
Hb_001671_060 |
0.1883373071 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 9 |
Hb_000545_170 |
0.1898603173 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 10 |
Hb_006018_030 |
0.192071129 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 11 |
Hb_001157_220 |
0.1969418929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000120_360 |
0.1982542382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_098315_030 |
0.1991083366 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 14 |
Hb_028285_030 |
0.2014058102 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 15 |
Hb_000235_010 |
0.2018218964 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 16 |
Hb_002000_110 |
0.2019124399 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 17 |
Hb_000614_100 |
0.2072085512 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] reductase [Populus trichocarpa] |
| 18 |
Hb_104010_010 |
0.2135610704 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |
| 19 |
Hb_001959_260 |
0.2139042013 |
- |
- |
- |
| 20 |
Hb_003050_070 |
0.216177191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |