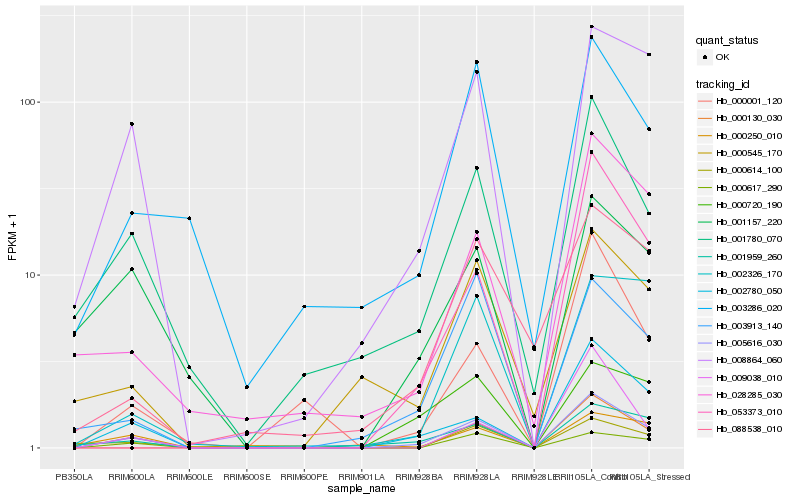
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005616_030 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 2 |
Hb_002780_050 |
0.1609888114 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105169583 [Sesamum indicum] |
| 3 |
Hb_008864_060 |
0.172994905 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 4 |
Hb_001780_070 |
0.1806469239 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 5 |
Hb_000617_290 |
0.1905649961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001959_260 |
0.1958178191 |
- |
- |
- |
| 7 |
Hb_000720_190 |
0.2065544228 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_000001_120 |
0.2119033967 |
- |
- |
hypothetical protein JCGZ_03063 [Jatropha curcas] |
| 9 |
Hb_028285_030 |
0.2179810703 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 10 |
Hb_003286_020 |
0.2195479158 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 11 |
Hb_000130_030 |
0.220650545 |
- |
- |
PREDICTED: small ubiquitin-related modifier 2-A-like [Eucalyptus grandis] |
| 12 |
Hb_003913_140 |
0.2211120001 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 13 |
Hb_000250_010 |
0.222037554 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_000545_170 |
0.2222058572 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 15 |
Hb_053373_010 |
0.2279762069 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 16 |
Hb_000614_100 |
0.2330794147 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] reductase [Populus trichocarpa] |
| 17 |
Hb_009038_010 |
0.2339483166 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 18 |
Hb_002326_170 |
0.2345767346 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_001157_220 |
0.2356276364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_088538_010 |
0.2366132463 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |