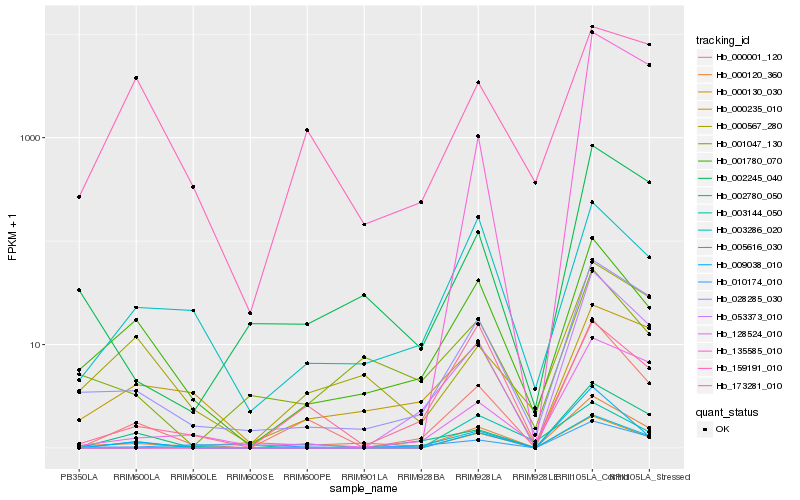
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_03063 [Jatropha curcas] |
| 2 |
Hb_009038_010 |
0.1813140745 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 3 |
Hb_002780_050 |
0.1979520054 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105169583 [Sesamum indicum] |
| 4 |
Hb_053373_010 |
0.1990365623 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_028285_030 |
0.2073551675 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 6 |
Hb_005616_030 |
0.2119033967 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 7 |
Hb_000120_360 |
0.2157917096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001780_070 |
0.2287570889 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 9 |
Hb_128524_010 |
0.2289505022 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000130_030 |
0.2325498707 |
- |
- |
PREDICTED: small ubiquitin-related modifier 2-A-like [Eucalyptus grandis] |
| 11 |
Hb_002245_040 |
0.252414026 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 12 |
Hb_000567_280 |
0.2643441815 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 13 |
Hb_010174_010 |
0.2646994416 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 14 |
Hb_135585_010 |
0.2701455462 |
- |
- |
- |
| 15 |
Hb_003286_020 |
0.2833875254 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 16 |
Hb_173281_010 |
0.2838673912 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_159191_010 |
0.2850264826 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 18 |
Hb_003144_050 |
0.2862128676 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 19 |
Hb_000235_010 |
0.2862772498 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 20 |
Hb_001047_130 |
0.290135197 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |