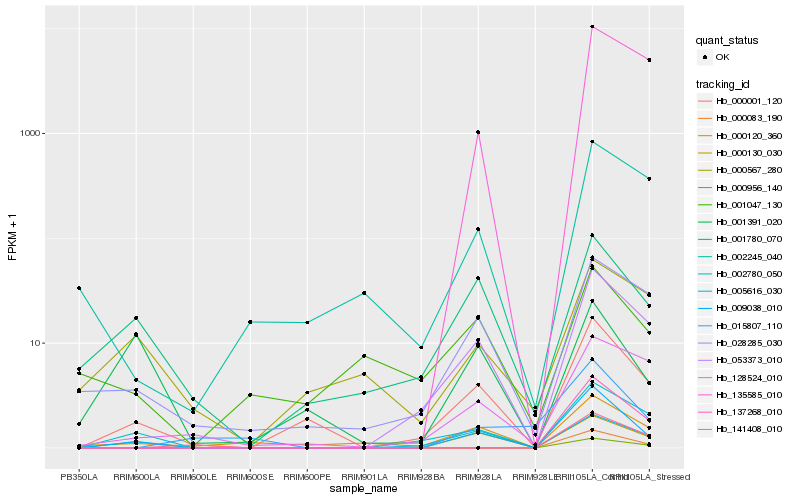
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009038_010 |
0.0 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 2 |
Hb_000001_120 |
0.1813140745 |
- |
- |
hypothetical protein JCGZ_03063 [Jatropha curcas] |
| 3 |
Hb_002780_050 |
0.2219568273 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105169583 [Sesamum indicum] |
| 4 |
Hb_053373_010 |
0.2242308163 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_000130_030 |
0.2334722938 |
- |
- |
PREDICTED: small ubiquitin-related modifier 2-A-like [Eucalyptus grandis] |
| 6 |
Hb_005616_030 |
0.2339483166 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 7 |
Hb_001780_070 |
0.2697867827 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 8 |
Hb_028285_030 |
0.2741441748 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 9 |
Hb_000120_360 |
0.2814009245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_135585_010 |
0.2877716241 |
- |
- |
- |
| 11 |
Hb_000083_190 |
0.2910772423 |
- |
- |
low affinity nitrate transporter, partial [Triticum aestivum] |
| 12 |
Hb_137268_010 |
0.3002928742 |
- |
- |
- |
| 13 |
Hb_015807_110 |
0.3008852148 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 14 |
Hb_128524_010 |
0.3017462918 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000956_140 |
0.3125701875 |
- |
- |
- |
| 16 |
Hb_002245_040 |
0.3139706134 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 17 |
Hb_001391_020 |
0.3226549975 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 18 |
Hb_000567_280 |
0.3301214445 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 19 |
Hb_001047_130 |
0.3420167684 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 20 |
Hb_141408_010 |
0.3429842536 |
- |
- |
PREDICTED: uncharacterized protein LOC105631382 [Jatropha curcas] |