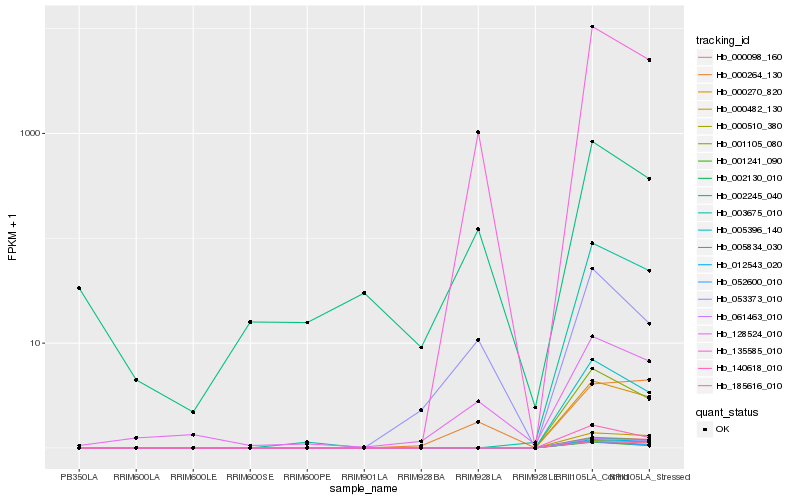
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135585_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_053373_010 |
0.1643015885 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_128524_010 |
0.1775034321 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005834_030 |
0.1793764649 |
- |
- |
- |
| 5 |
Hb_001105_080 |
0.181049754 |
- |
- |
- |
| 6 |
Hb_005396_140 |
0.1820808421 |
- |
- |
- |
| 7 |
Hb_140618_010 |
0.1825797551 |
- |
- |
- |
| 8 |
Hb_001241_090 |
0.1825864055 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 9 |
Hb_052600_010 |
0.1826112261 |
- |
- |
- |
| 10 |
Hb_003675_010 |
0.1826234049 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 11 |
Hb_000270_820 |
0.1850261817 |
- |
- |
- |
| 12 |
Hb_000264_130 |
0.189888681 |
- |
- |
PREDICTED: uncharacterized protein LOC103870402 [Brassica rapa] |
| 13 |
Hb_002245_040 |
0.2002617287 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 14 |
Hb_000510_380 |
0.2043685017 |
- |
- |
- |
| 15 |
Hb_000482_130 |
0.2043708341 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_012543_020 |
0.2043729129 |
- |
- |
- |
| 17 |
Hb_185616_010 |
0.2043762474 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 18 |
Hb_061463_010 |
0.2043940331 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 19 |
Hb_002130_010 |
0.204400179 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 20 |
Hb_000098_160 |
0.2044309384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |