| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
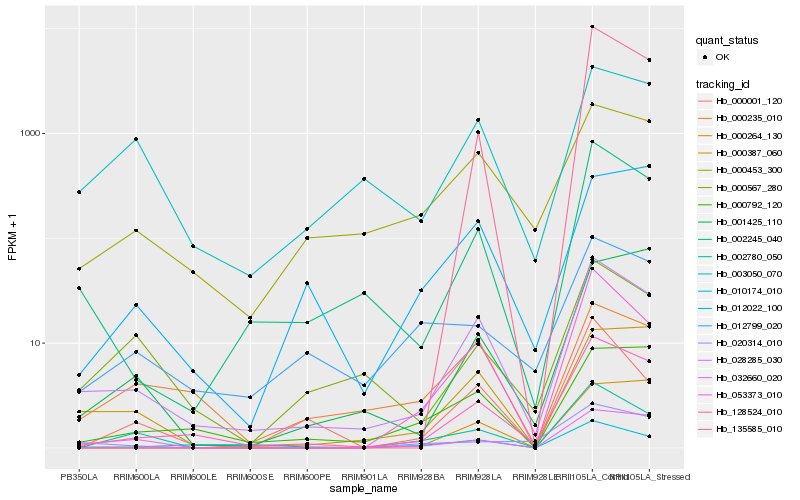
Hb_128524_010 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028285_030 |
0.1259993004 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 3 |
Hb_002245_040 |
0.1692086568 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 4 |
Hb_000792_120 |
0.17613655 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_135585_010 |
0.1775034321 |
- |
- |
- |
| 6 |
Hb_053373_010 |
0.1976567512 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000453_300 |
0.2049294116 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 8 |
Hb_000264_130 |
0.2100524119 |
- |
- |
PREDICTED: uncharacterized protein LOC103870402 [Brassica rapa] |
| 9 |
Hb_012022_100 |
0.2144000847 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 10 |
Hb_002780_050 |
0.2150846561 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105169583 [Sesamum indicum] |
| 11 |
Hb_000387_060 |
0.2159694438 |
- |
- |
- |
| 12 |
Hb_010174_010 |
0.2162176346 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 13 |
Hb_001425_110 |
0.2180669898 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 14 |
Hb_012799_020 |
0.2199174873 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_032660_020 |
0.2206986403 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 16 |
Hb_000235_010 |
0.2288977818 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 17 |
Hb_000001_120 |
0.2289505022 |
- |
- |
hypothetical protein JCGZ_03063 [Jatropha curcas] |
| 18 |
Hb_000567_280 |
0.2313271747 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 19 |
Hb_020314_010 |
0.2336706508 |
- |
- |
- |
| 20 |
Hb_003050_070 |
0.2368662789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |