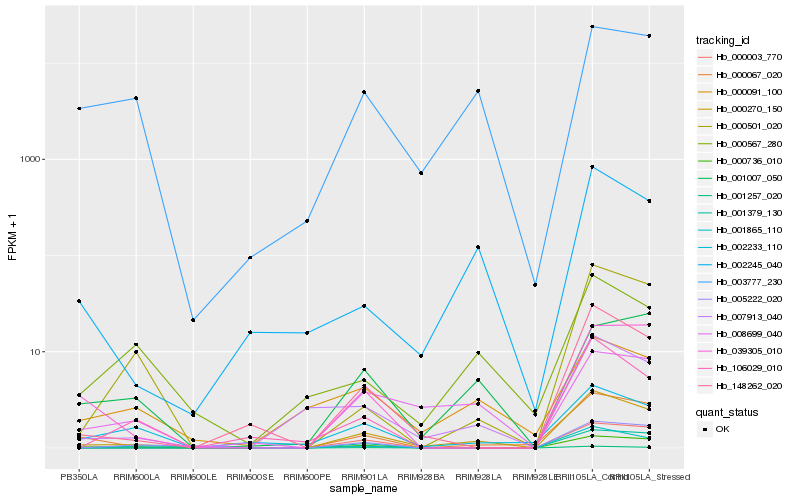
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_150 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_148262_020 |
0.1764686923 |
- |
- |
- |
| 3 |
Hb_000067_020 |
0.1794535859 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 4 |
Hb_007913_040 |
0.1857753098 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 5 |
Hb_002245_040 |
0.1985750002 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 6 |
Hb_001257_020 |
0.205501639 |
- |
- |
- |
| 7 |
Hb_000736_010 |
0.2085451995 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 8 |
Hb_001379_130 |
0.2133155614 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 9 |
Hb_005222_020 |
0.2143229816 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_003777_230 |
0.2164069008 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 11 |
Hb_000003_770 |
0.2183528226 |
- |
- |
- |
| 12 |
Hb_000501_020 |
0.2247305265 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 13 |
Hb_008699_040 |
0.2251486551 |
- |
- |
- |
| 14 |
Hb_001865_110 |
0.2260400848 |
- |
- |
- |
| 15 |
Hb_039305_010 |
0.2287816375 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 16 |
Hb_001007_050 |
0.2317850164 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 17 |
Hb_000567_280 |
0.2324477623 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 18 |
Hb_106029_010 |
0.235029382 |
- |
- |
Transcription factor ICE1, putative [Ricinus communis] |
| 19 |
Hb_000091_100 |
0.2361759972 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_002233_110 |
0.2374184052 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |