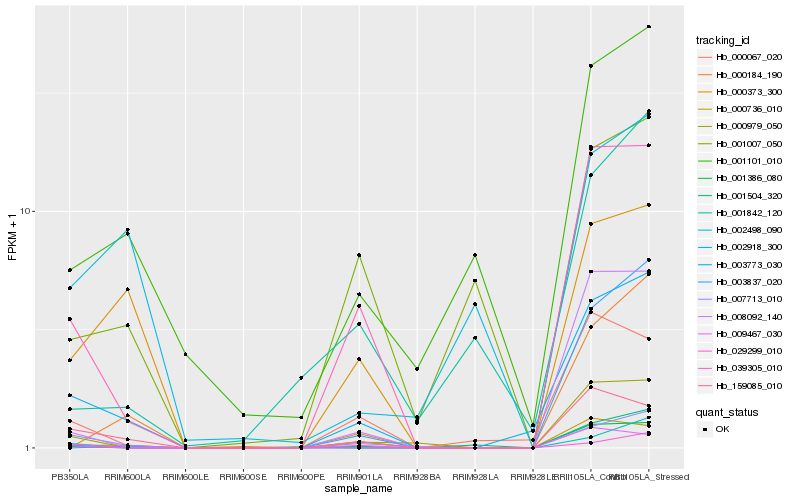
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003773_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_039305_010 |
0.1665388045 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 3 |
Hb_007713_010 |
0.187649586 |
- |
- |
hypothetical protein JCGZ_18011 [Jatropha curcas] |
| 4 |
Hb_159085_010 |
0.2021517369 |
- |
- |
- |
| 5 |
Hb_001504_320 |
0.2038436691 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 6 |
Hb_000067_020 |
0.2038877473 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 7 |
Hb_000979_050 |
0.2080449223 |
- |
- |
- |
| 8 |
Hb_000373_300 |
0.217877435 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 9 |
Hb_008092_140 |
0.2207542017 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 10 |
Hb_001101_010 |
0.2211574596 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 11 |
Hb_000736_010 |
0.2292281361 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 12 |
Hb_000184_190 |
0.2322089462 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 13 |
Hb_029299_010 |
0.2453606169 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 14 |
Hb_002498_090 |
0.2546171131 |
- |
- |
- |
| 15 |
Hb_001386_080 |
0.2566104038 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 16 |
Hb_003837_020 |
0.2572148939 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 17 |
Hb_009467_030 |
0.2634572268 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 18 |
Hb_001842_120 |
0.2640124552 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 19 |
Hb_001007_050 |
0.2691261779 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 20 |
Hb_002918_300 |
0.2700037522 |
- |
- |
- |