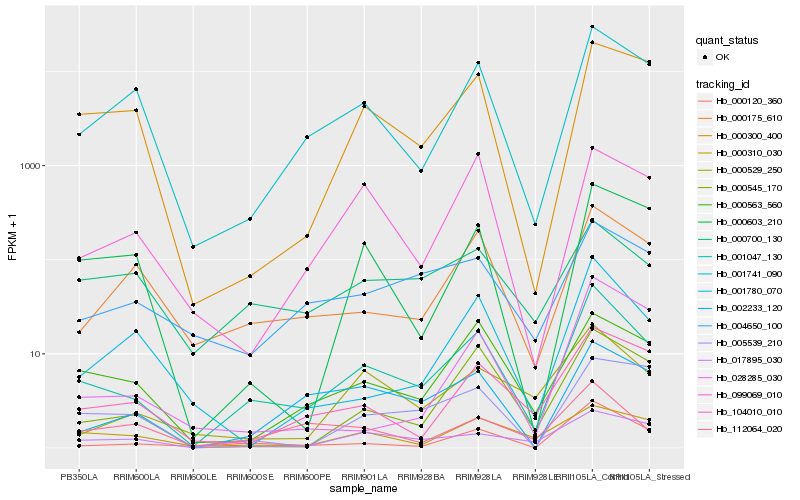
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001047_130 |
0.0 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 2 |
Hb_000120_360 |
0.1571266089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001741_090 |
0.1876021665 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 4 |
Hb_000545_170 |
0.2057277387 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 5 |
Hb_000175_610 |
0.209038124 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 6 |
Hb_104010_010 |
0.2094514906 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |
| 7 |
Hb_000563_560 |
0.2120993463 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 8 |
Hb_000529_250 |
0.2133503387 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002233_120 |
0.2152703571 |
- |
- |
- |
| 10 |
Hb_017895_030 |
0.2176665548 |
transcription factor |
TF Family: C2H2 |
- |
| 11 |
Hb_001780_070 |
0.2178456906 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 12 |
Hb_000300_400 |
0.2189856509 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 13 |
Hb_004650_100 |
0.2221211559 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000603_210 |
0.2289809348 |
- |
- |
hypothetical protein POPTR_0006s23660g [Populus trichocarpa] |
| 15 |
Hb_000700_130 |
0.2349696968 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 16 |
Hb_099069_010 |
0.2357576604 |
- |
- |
S-adenosyl-methionine-sterol-C- methyltransferase [Ricinus communis] |
| 17 |
Hb_000310_030 |
0.2369298289 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 18 |
Hb_028285_030 |
0.2375561195 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 19 |
Hb_112064_020 |
0.2379558689 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 20 |
Hb_005539_210 |
0.2397078117 |
- |
- |
- |