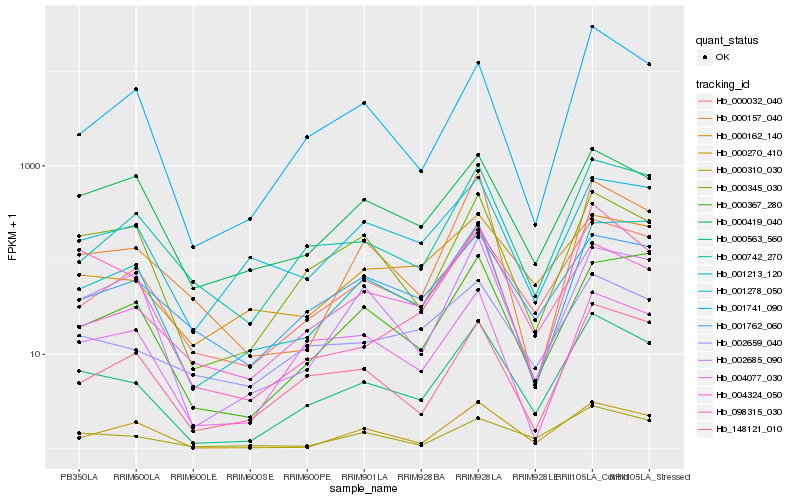
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_560 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 2 |
Hb_000032_040 |
0.1329458174 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 3 |
Hb_000742_270 |
0.1462648096 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 4 |
Hb_000310_030 |
0.148392451 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 5 |
Hb_000345_030 |
0.1496153425 |
- |
- |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 6 |
Hb_002659_040 |
0.1497817324 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_148121_010 |
0.152675377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000270_410 |
0.1592020164 |
- |
- |
- |
| 9 |
Hb_001762_060 |
0.1607851152 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 10 |
Hb_000157_040 |
0.1611960007 |
- |
- |
PREDICTED: cycloartenol-C-24-methyltransferase [Jatropha curcas] |
| 11 |
Hb_004324_050 |
0.1612760632 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 12 |
Hb_001213_120 |
0.162287088 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 13 |
Hb_000162_140 |
0.1647384872 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2 [Jatropha curcas] |
| 14 |
Hb_000419_040 |
0.1649393209 |
- |
- |
chorismate synthase [Hevea brasiliensis] |
| 15 |
Hb_002685_090 |
0.165818396 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 16 |
Hb_001278_050 |
0.166053898 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 17 |
Hb_098315_030 |
0.1663130492 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 18 |
Hb_004077_030 |
0.1671265054 |
- |
- |
hypothetical protein EUGRSUZ_K01571 [Eucalyptus grandis] |
| 19 |
Hb_001741_090 |
0.1708511995 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 20 |
Hb_000367_280 |
0.1714964618 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |