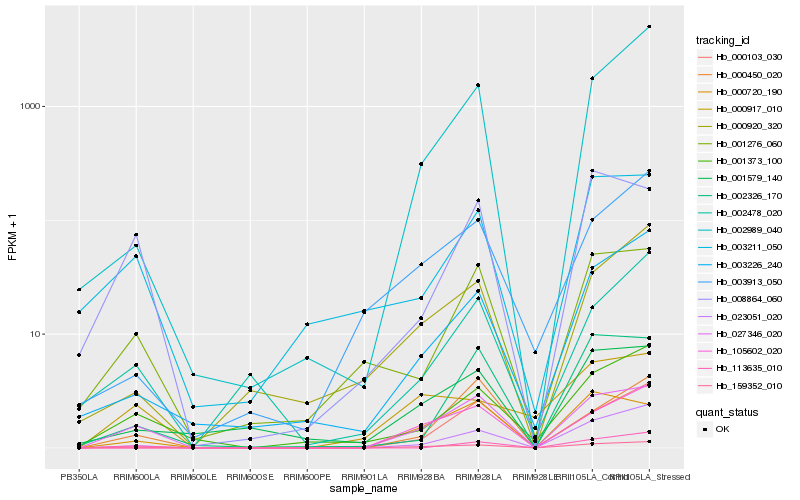
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159352_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_12393 [Jatropha curcas] |
| 2 |
Hb_027346_020 |
0.0898133481 |
- |
- |
- |
| 3 |
Hb_023051_020 |
0.1604592826 |
- |
- |
- |
| 4 |
Hb_000720_190 |
0.1814527456 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_001373_100 |
0.1830961973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003226_240 |
0.1921780617 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_105602_020 |
0.1929118269 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 8 |
Hb_002989_040 |
0.1957840805 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 9 |
Hb_001579_140 |
0.201539406 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000920_320 |
0.2056022173 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 11 |
Hb_113635_010 |
0.2121673137 |
- |
- |
- |
| 12 |
Hb_001276_060 |
0.2123133043 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 13 |
Hb_003913_050 |
0.2165455306 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 14 |
Hb_003211_050 |
0.2171207335 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000450_020 |
0.2192080698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008864_060 |
0.2195143874 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 17 |
Hb_002478_020 |
0.221571715 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 18 |
Hb_002326_170 |
0.2216274475 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_000917_010 |
0.2219730459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000103_030 |
0.2223585869 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |