| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
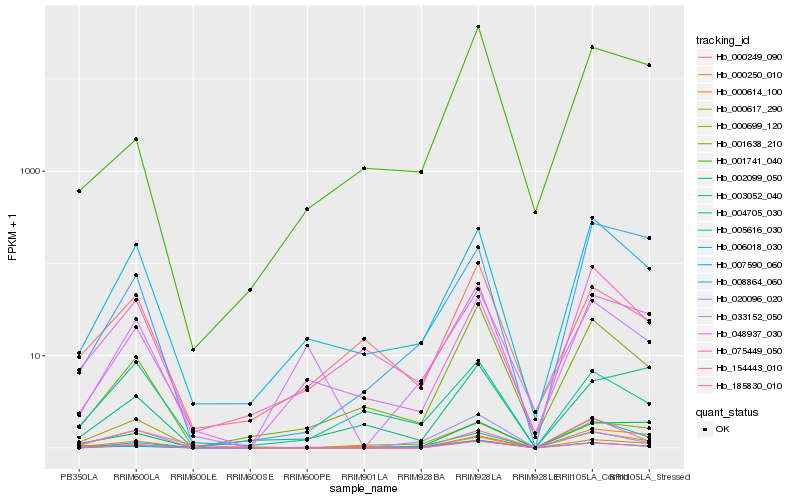
Hb_154443_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 2 |
Hb_000617_290 |
0.1456338998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001638_210 |
0.1976220488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006018_030 |
0.2062923698 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 5 |
Hb_048937_030 |
0.2333721925 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 6 |
Hb_002099_050 |
0.2441224122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_020096_020 |
0.250685891 |
- |
- |
- |
| 8 |
Hb_033152_050 |
0.2636920775 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 9 |
Hb_000249_090 |
0.2643229279 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 10 |
Hb_185830_010 |
0.2646276335 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1 [Jatropha curcas] |
| 11 |
Hb_000614_100 |
0.2718697305 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] reductase [Populus trichocarpa] |
| 12 |
Hb_000699_120 |
0.2727807833 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 13 |
Hb_075449_050 |
0.2748047269 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 14 |
Hb_008864_060 |
0.2754614855 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 15 |
Hb_001741_040 |
0.2765346746 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 16 |
Hb_005616_030 |
0.2833095026 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Eucalyptus grandis] |
| 17 |
Hb_000250_010 |
0.2875515059 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 18 |
Hb_004705_030 |
0.2904676725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003052_040 |
0.2913656222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007590_060 |
0.2920008429 |
- |
- |
- |