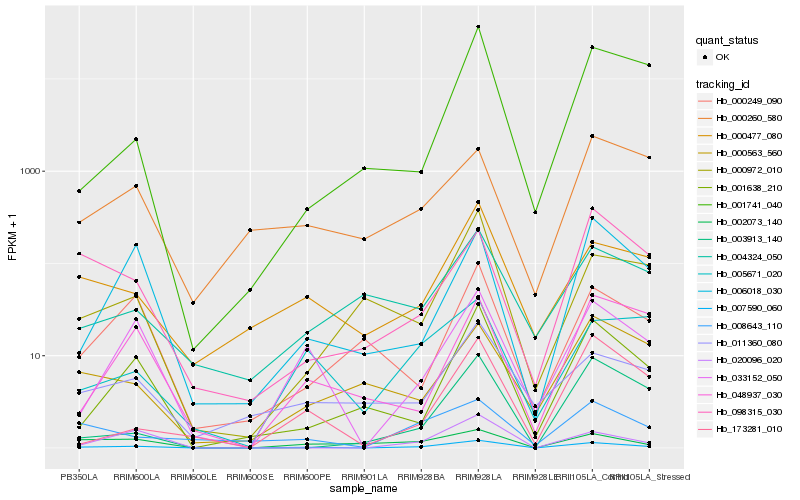
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007590_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_033152_050 |
0.1962662304 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 3 |
Hb_001638_210 |
0.2139627832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_020096_020 |
0.221029216 |
- |
- |
- |
| 5 |
Hb_000477_080 |
0.2300340941 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 6 |
Hb_006018_030 |
0.2393763472 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 7 |
Hb_003913_140 |
0.2413142596 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 8 |
Hb_173281_010 |
0.2446902842 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_008643_110 |
0.2475447343 |
- |
- |
BnaCnng39750D [Brassica napus] |
| 10 |
Hb_002073_140 |
0.2480560854 |
- |
- |
- |
| 11 |
Hb_011360_080 |
0.2510242502 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000972_010 |
0.2522052287 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 13 |
Hb_005671_020 |
0.252562686 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 14 |
Hb_048937_030 |
0.2541054019 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 15 |
Hb_000249_090 |
0.2549793241 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 16 |
Hb_001741_040 |
0.2586719695 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 17 |
Hb_098315_030 |
0.261150056 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 18 |
Hb_004324_050 |
0.2617272793 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 19 |
Hb_000563_560 |
0.2632200928 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 20 |
Hb_000260_580 |
0.2651282806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |