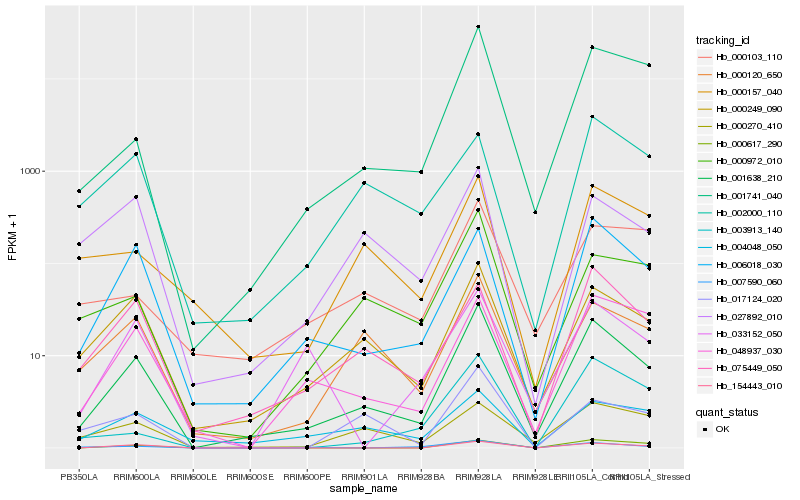
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000249_090 |
0.1590638401 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 3 |
Hb_006018_030 |
0.1724796259 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 4 |
Hb_001741_040 |
0.1857954402 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 5 |
Hb_000120_650 |
0.1866309036 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 6 |
Hb_075449_050 |
0.1924659237 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 7 |
Hb_048937_030 |
0.1934910902 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 8 |
Hb_154443_010 |
0.1976220488 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 9 |
Hb_000972_010 |
0.2006014726 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 10 |
Hb_000157_040 |
0.2058626834 |
- |
- |
PREDICTED: cycloartenol-C-24-methyltransferase [Jatropha curcas] |
| 11 |
Hb_007590_060 |
0.2139627832 |
- |
- |
- |
| 12 |
Hb_027892_010 |
0.2151439549 |
transcription factor |
TF Family: bHLH |
LMYC1 [Hevea brasiliensis] |
| 13 |
Hb_017124_020 |
0.2227190614 |
- |
- |
hypothetical protein PRUPE_ppa015539mg [Prunus persica] |
| 14 |
Hb_003913_140 |
0.2245586069 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 15 |
Hb_000103_110 |
0.225022639 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000270_410 |
0.2263035207 |
- |
- |
- |
| 17 |
Hb_004048_050 |
0.2270438652 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 18 |
Hb_000617_290 |
0.2290271424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_033152_050 |
0.2305832701 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 20 |
Hb_002000_110 |
0.2330732038 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |