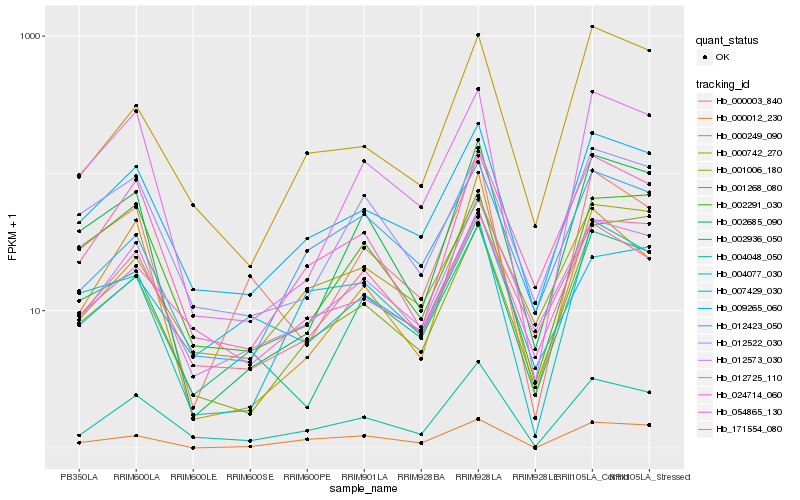
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004048_050 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 2 |
Hb_009265_060 |
0.0954515254 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 3 |
Hb_012573_030 |
0.1099109034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_054865_130 |
0.1236007904 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_024714_060 |
0.127195626 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_171554_080 |
0.1344025229 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_012725_110 |
0.144194941 |
- |
- |
hypothetical protein JCGZ_09397 [Jatropha curcas] |
| 8 |
Hb_000742_270 |
0.1471175216 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 9 |
Hb_001006_180 |
0.1483651556 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 10 |
Hb_000249_090 |
0.1514051688 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 11 |
Hb_000003_840 |
0.1540187275 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |
| 12 |
Hb_007429_030 |
0.1547064625 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002685_090 |
0.155639624 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 14 |
Hb_002936_050 |
0.1567303086 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012423_050 |
0.1588588577 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 16 |
Hb_000012_230 |
0.1592283807 |
- |
- |
PREDICTED: uncharacterized protein LOC105629874 [Jatropha curcas] |
| 17 |
Hb_004077_030 |
0.16082495 |
- |
- |
hypothetical protein EUGRSUZ_K01571 [Eucalyptus grandis] |
| 18 |
Hb_002291_030 |
0.1631768519 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 19 |
Hb_012522_030 |
0.1638895967 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001268_080 |
0.1640247877 |
- |
- |
catalytic, putative [Ricinus communis] |