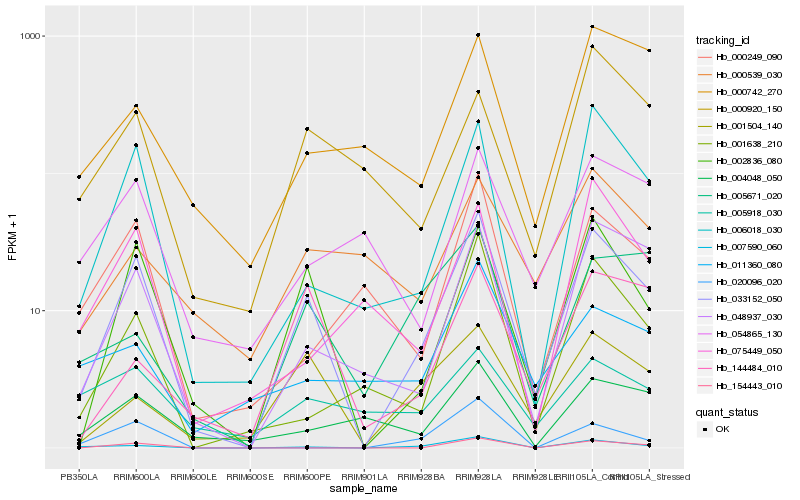
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033152_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_04890 [Jatropha curcas] |
| 2 |
Hb_002836_080 |
0.1495380247 |
- |
- |
- |
| 3 |
Hb_006018_030 |
0.1958774678 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 4 |
Hb_007590_060 |
0.1962662304 |
- |
- |
- |
| 5 |
Hb_048937_030 |
0.1985195969 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 6 |
Hb_001504_140 |
0.225683813 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 7 |
Hb_001638_210 |
0.2305832701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000249_090 |
0.2451191133 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 9 |
Hb_144484_010 |
0.2553515517 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 10 |
Hb_004048_050 |
0.2557009356 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 11 |
Hb_000920_150 |
0.2569988606 |
- |
- |
annexin [Manihot esculenta] |
| 12 |
Hb_054865_130 |
0.2573469793 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_075449_050 |
0.2582236679 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 14 |
Hb_000742_270 |
0.2620154413 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 15 |
Hb_154443_010 |
0.2636920775 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 16 |
Hb_005671_020 |
0.2653792483 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 17 |
Hb_011360_080 |
0.2664710006 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_020096_020 |
0.2672076432 |
- |
- |
- |
| 19 |
Hb_000539_030 |
0.2696510095 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 20 |
Hb_005918_030 |
0.2709132028 |
- |
- |
hypothetical protein JCGZ_24769 [Jatropha curcas] |