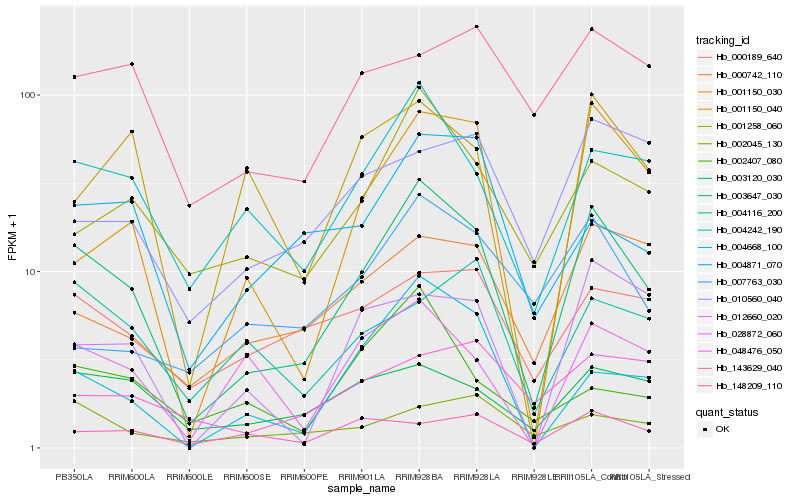
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003647_030 |
0.0 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 2 |
Hb_004242_190 |
0.1771808798 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 3 |
Hb_001258_060 |
0.1947572809 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 4 |
Hb_001150_030 |
0.2000211203 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 5 |
Hb_028872_060 |
0.2088355409 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 6 |
Hb_004668_100 |
0.2123812186 |
- |
- |
- |
| 7 |
Hb_003120_030 |
0.2193302938 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 8 |
Hb_004871_070 |
0.2211860937 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 9 |
Hb_000742_110 |
0.2215834926 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 10 |
Hb_000189_640 |
0.2260843838 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 11 |
Hb_012660_020 |
0.2293397358 |
- |
- |
- |
| 12 |
Hb_001150_040 |
0.2294845936 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 13 |
Hb_002045_130 |
0.2346304982 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 14 |
Hb_007763_030 |
0.238307258 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 15 |
Hb_143629_040 |
0.2392201178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010560_040 |
0.2412637559 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 17 |
Hb_148209_110 |
0.2427226817 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 18 |
Hb_004116_200 |
0.2460339681 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 19 |
Hb_002407_080 |
0.2496547693 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 20 |
Hb_048476_050 |
0.2497192668 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |