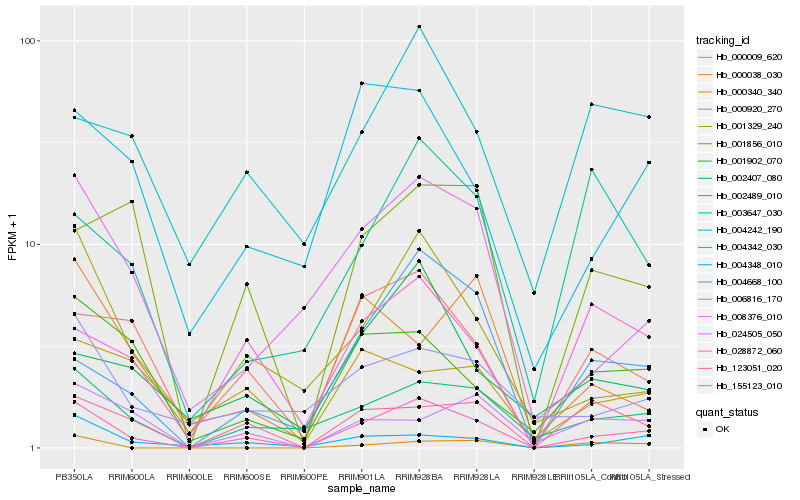
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123051_020 |
0.0 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 2 |
Hb_002489_010 |
0.2017989121 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 3 |
Hb_001329_240 |
0.2105130251 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 4 |
Hb_000009_620 |
0.2170580239 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_024505_050 |
0.2245311871 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 6 |
Hb_004348_010 |
0.2292389315 |
- |
- |
hypothetical protein POPTR_0077s002302g, partial [Populus trichocarpa] |
| 7 |
Hb_155123_010 |
0.233365764 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_004668_100 |
0.2366811941 |
- |
- |
- |
| 9 |
Hb_001856_010 |
0.2450231868 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001902_070 |
0.2450780997 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_004342_030 |
0.247352123 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 12 |
Hb_006816_170 |
0.2506865411 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 13 |
Hb_028872_060 |
0.2528066019 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 14 |
Hb_008376_010 |
0.2649700925 |
- |
- |
- |
| 15 |
Hb_002407_080 |
0.2659667986 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 16 |
Hb_004242_190 |
0.2689366309 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_000920_270 |
0.2724547202 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 18 |
Hb_003647_030 |
0.278891008 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 19 |
Hb_000340_340 |
0.2857383512 |
- |
- |
hypothetical protein POPTR_0014s13260g [Populus trichocarpa] |
| 20 |
Hb_000038_030 |
0.2917729536 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |