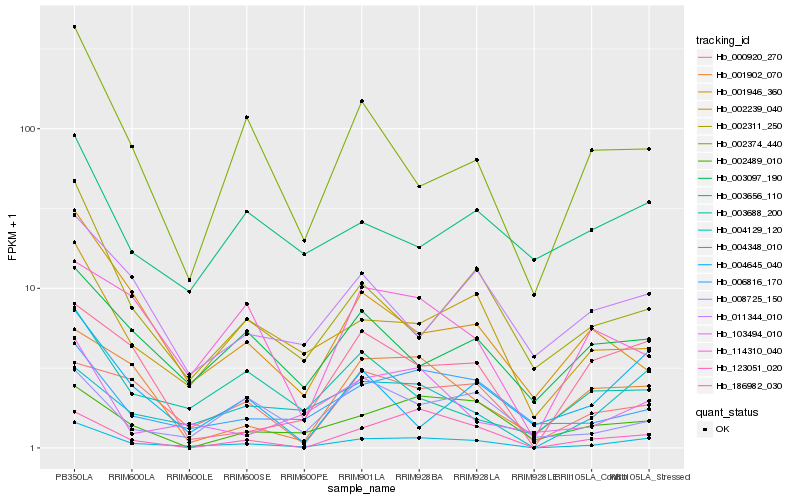
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004348_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0077s002302g, partial [Populus trichocarpa] |
| 2 |
Hb_002239_040 |
0.2006206261 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_006816_170 |
0.201904844 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 4 |
Hb_002311_250 |
0.212727274 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 5 |
Hb_004129_120 |
0.2161300785 |
- |
- |
- |
| 6 |
Hb_002374_440 |
0.2179336492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001902_070 |
0.2240731116 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_186982_030 |
0.2243173613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000920_270 |
0.227782016 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 10 |
Hb_123051_020 |
0.2292389315 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 11 |
Hb_004645_040 |
0.2292668971 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 12 |
Hb_003097_190 |
0.2299514351 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 13 |
Hb_103494_010 |
0.231763017 |
- |
- |
- |
| 14 |
Hb_002489_010 |
0.2335573874 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 15 |
Hb_011344_010 |
0.2370877999 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 16 |
Hb_001946_360 |
0.2377411425 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 17 |
Hb_008725_150 |
0.2426713825 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 18 |
Hb_003688_200 |
0.2429128131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_114310_040 |
0.2440988442 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 20 |
Hb_003656_110 |
0.2464153274 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |