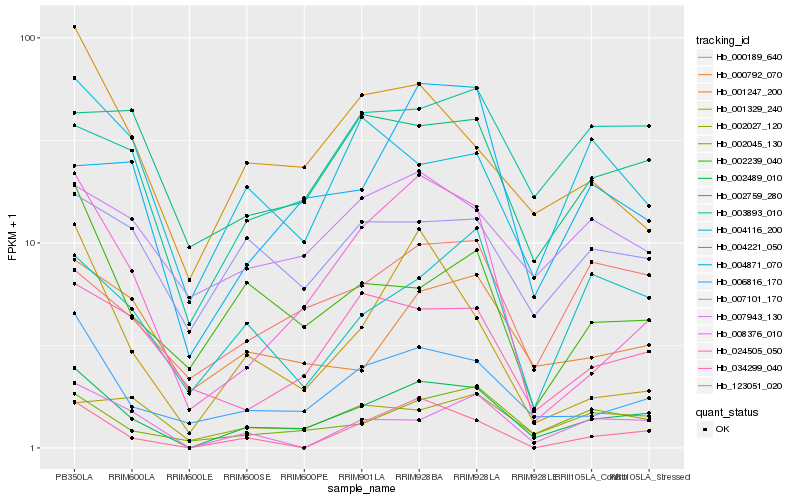
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002489_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 2 |
Hb_006816_170 |
0.1497579221 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 3 |
Hb_002045_130 |
0.166377474 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 4 |
Hb_024505_050 |
0.1811068672 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 5 |
Hb_000792_070 |
0.1959553622 |
- |
- |
Ataxin-3, putative [Ricinus communis] |
| 6 |
Hb_004116_200 |
0.1968945611 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 7 |
Hb_003893_010 |
0.1976357859 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 8 |
Hb_001247_200 |
0.197803671 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 9 |
Hb_034299_040 |
0.200000054 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 10 |
Hb_000189_640 |
0.2013683715 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 11 |
Hb_002239_040 |
0.2015793566 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_123051_020 |
0.2017989121 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 13 |
Hb_007101_170 |
0.2034084355 |
- |
- |
hypothetical protein POPTR_0010s10490g [Populus trichocarpa] |
| 14 |
Hb_007943_130 |
0.2067266424 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 15 |
Hb_008376_010 |
0.2098959549 |
- |
- |
- |
| 16 |
Hb_002759_280 |
0.2111465854 |
- |
- |
PREDICTED: uncharacterized protein LOC105649824 [Jatropha curcas] |
| 17 |
Hb_004221_050 |
0.2137030708 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 18 |
Hb_001329_240 |
0.2137664704 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 19 |
Hb_002027_120 |
0.2156278146 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 20 |
Hb_004871_070 |
0.2156387013 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |