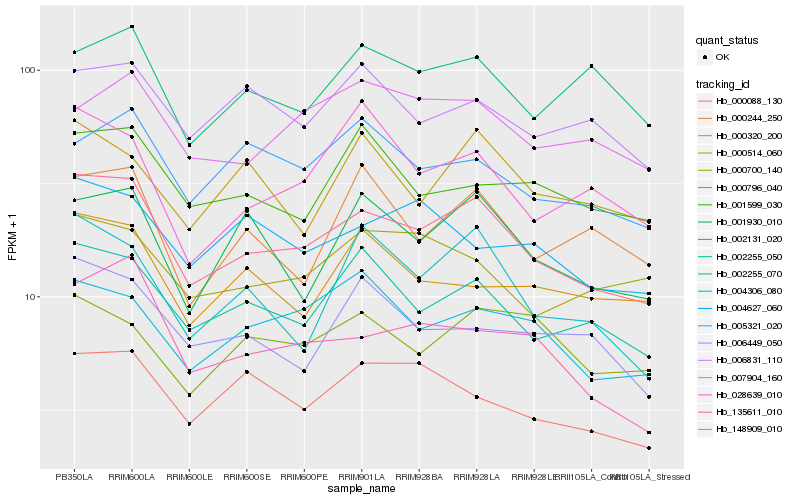
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148909_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002255_050 |
0.0827049832 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_004306_080 |
0.0999451343 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 4 |
Hb_135611_010 |
0.1026248983 |
- |
- |
PREDICTED: uncharacterized protein LOC105142085 [Populus euphratica] |
| 5 |
Hb_001930_010 |
0.1039948771 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 6 |
Hb_002255_070 |
0.1058553751 |
- |
- |
PREDICTED: isoleucine--tRNA ligase, cytoplasmic-like [Camelina sativa] |
| 7 |
Hb_000088_130 |
0.1058717891 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 8 |
Hb_005321_020 |
0.1070792358 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 9 |
Hb_006831_110 |
0.108018749 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 10 |
Hb_000700_140 |
0.1087792408 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 11 |
Hb_001599_030 |
0.1096700056 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_006449_050 |
0.1104075981 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 13 |
Hb_004627_060 |
0.1113482891 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 14 |
Hb_002131_020 |
0.1114347396 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 15 |
Hb_000320_200 |
0.1115395856 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 16 |
Hb_028639_010 |
0.111581514 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_007904_160 |
0.1116349352 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 18 |
Hb_000514_060 |
0.1122409227 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG isoform X1 [Jatropha curcas] |
| 19 |
Hb_000796_040 |
0.1123263687 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000244_250 |
0.1133932227 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |