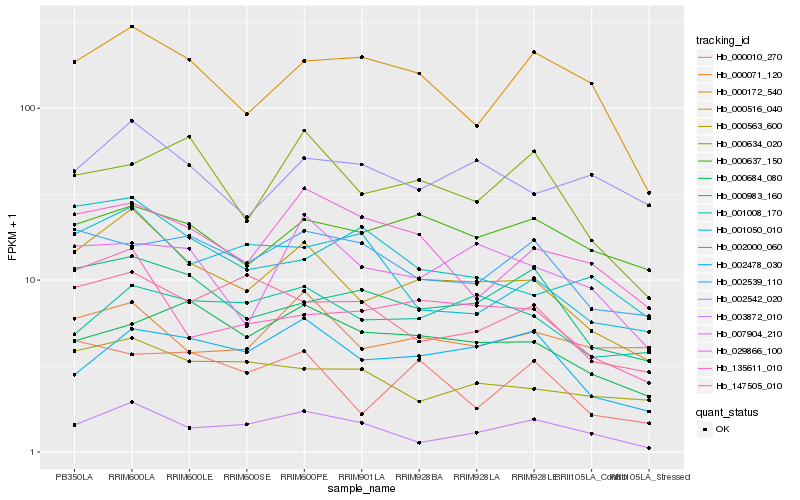
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000516_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 2 |
Hb_135611_010 |
0.1272067267 |
- |
- |
PREDICTED: uncharacterized protein LOC105142085 [Populus euphratica] |
| 3 |
Hb_000983_160 |
0.1401136794 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 4 |
Hb_000563_600 |
0.1488807277 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 5 |
Hb_000172_540 |
0.1555508214 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 6 |
Hb_002478_030 |
0.1555648665 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 7 |
Hb_029866_100 |
0.155804432 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 8 |
Hb_000071_120 |
0.1569702962 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 9 |
Hb_003872_010 |
0.1572721812 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 10 |
Hb_007904_210 |
0.1582245044 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 11 |
Hb_001008_170 |
0.1582511196 |
transcription factor |
TF Family: mTERF |
RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_002542_020 |
0.1599154607 |
- |
- |
PREDICTED: uncharacterized protein LOC105634003 isoform X1 [Jatropha curcas] |
| 13 |
Hb_147505_010 |
0.1610961952 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 14 |
Hb_000637_150 |
0.1619564874 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 15 |
Hb_000684_080 |
0.1623381593 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 16 |
Hb_000010_270 |
0.1632735828 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 17 |
Hb_001050_010 |
0.1634981481 |
- |
- |
Advillin [Gossypium arboreum] |
| 18 |
Hb_002000_060 |
0.1666092736 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 19 |
Hb_000634_020 |
0.1675945588 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 20 |
Hb_002539_110 |
0.1677895286 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |