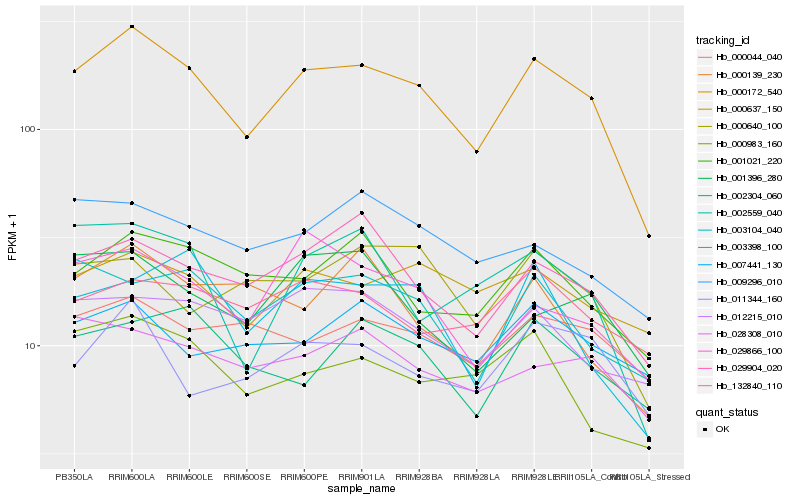
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_540 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 2 |
Hb_029904_020 |
0.1059636216 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 3 |
Hb_001021_220 |
0.1128966062 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003104_040 |
0.1152679925 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 5 |
Hb_029866_100 |
0.1173521947 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 6 |
Hb_000983_160 |
0.1197330474 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 7 |
Hb_003398_100 |
0.1199804777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012215_010 |
0.121395611 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 9 |
Hb_009296_010 |
0.1218193052 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 10 |
Hb_028308_010 |
0.1222163322 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000637_150 |
0.1228145378 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 12 |
Hb_001396_280 |
0.1229976698 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 13 |
Hb_002559_040 |
0.1233580265 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 5 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002304_060 |
0.1254876078 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 15 |
Hb_011344_160 |
0.1274237598 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 16 |
Hb_000044_040 |
0.1277607429 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 17 |
Hb_000640_100 |
0.1280896245 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_132840_110 |
0.1296445471 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000139_230 |
0.1296522125 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 20 |
Hb_007441_130 |
0.1309229485 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |