| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
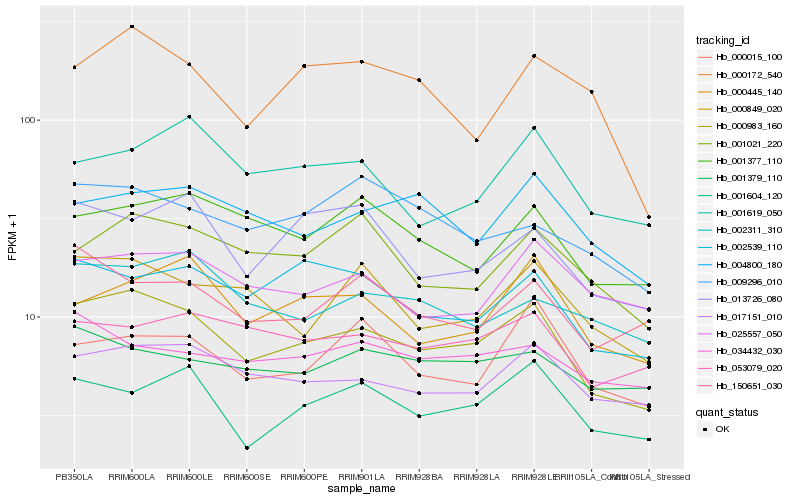
Hb_000983_160 |
0.0 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 2 |
Hb_017151_010 |
0.0949270297 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 3 |
Hb_000849_020 |
0.1045414568 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001604_120 |
0.1076367552 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002539_110 |
0.1076818272 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 6 |
Hb_000445_140 |
0.1123649419 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 7 |
Hb_001021_220 |
0.1129201891 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001377_110 |
0.1140929048 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 9 |
Hb_053079_020 |
0.1144632929 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 10 |
Hb_004800_180 |
0.1147960557 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 11 |
Hb_013726_080 |
0.1158389789 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 12 |
Hb_002311_310 |
0.1171984671 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 13 |
Hb_150651_030 |
0.1178126128 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 14 |
Hb_025557_050 |
0.1181212504 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 15 |
Hb_000172_540 |
0.1197330474 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 16 |
Hb_001379_110 |
0.1197488437 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 17 |
Hb_009296_010 |
0.119950257 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 18 |
Hb_000015_100 |
0.1201534509 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001619_050 |
0.1208329842 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 20 |
Hb_034432_030 |
0.1211451098 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |