| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
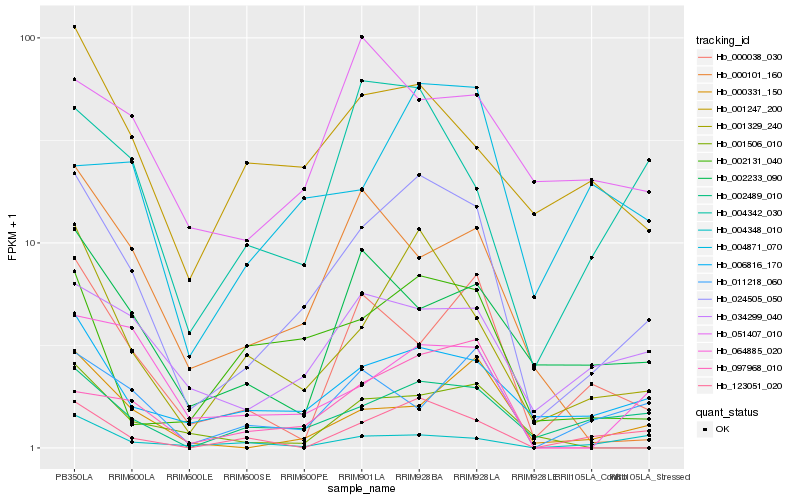
Hb_024505_050 |
0.0 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 2 |
Hb_001329_240 |
0.1707248255 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 3 |
Hb_006816_170 |
0.1795850158 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 4 |
Hb_002489_010 |
0.1811068672 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 5 |
Hb_034299_040 |
0.209019161 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_002131_040 |
0.2187220713 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 7 |
Hb_004342_030 |
0.2202492551 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 8 |
Hb_123051_020 |
0.2245311871 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 9 |
Hb_001247_200 |
0.2265521939 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 10 |
Hb_064885_020 |
0.2364023143 |
- |
- |
- |
| 11 |
Hb_000331_150 |
0.2388801248 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_004871_070 |
0.2448050174 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 13 |
Hb_002233_090 |
0.2452263096 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_000038_030 |
0.2467486707 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 15 |
Hb_001506_010 |
0.2516556299 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 16 |
Hb_097968_010 |
0.2569735768 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 17 |
Hb_011218_060 |
0.2569998848 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 18 |
Hb_000101_160 |
0.2592943723 |
- |
- |
PREDICTED: uncharacterized protein LOC105133607 isoform X1 [Populus euphratica] |
| 19 |
Hb_051407_010 |
0.2604807496 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_004348_010 |
0.2625799 |
- |
- |
hypothetical protein POPTR_0077s002302g, partial [Populus trichocarpa] |