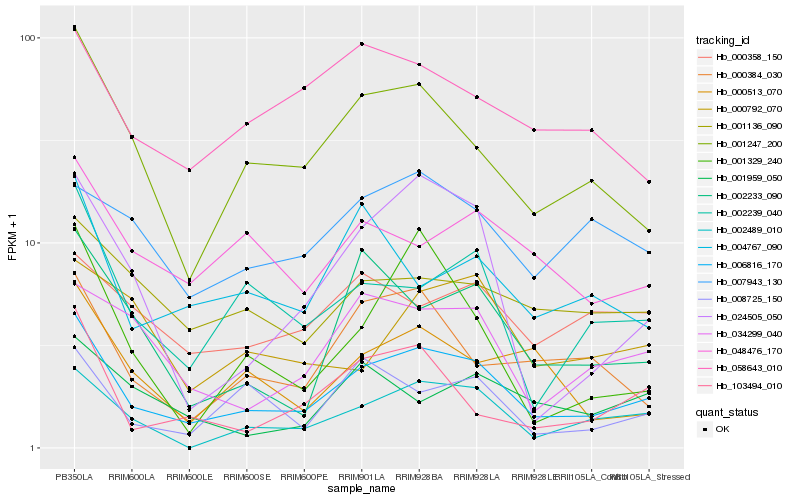
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_170 |
0.0 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 2 |
Hb_001247_200 |
0.1412357462 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 3 |
Hb_002489_010 |
0.1497579221 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 4 |
Hb_002239_040 |
0.1637702384 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_034299_040 |
0.1741713751 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_000384_030 |
0.1765286234 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_002233_090 |
0.1794032052 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_024505_050 |
0.1795850158 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 9 |
Hb_048476_170 |
0.1807473236 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |
| 10 |
Hb_001136_090 |
0.1819686164 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 11 |
Hb_000513_070 |
0.1857154662 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 12 |
Hb_058643_010 |
0.1861880155 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 13 |
Hb_004767_090 |
0.1869154263 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 14 |
Hb_008725_150 |
0.1876467162 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_103494_010 |
0.1879089368 |
- |
- |
- |
| 16 |
Hb_000358_150 |
0.1903517877 |
- |
- |
hypothetical protein VITISV_012309 [Vitis vinifera] |
| 17 |
Hb_007943_130 |
0.1953903699 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 18 |
Hb_000792_070 |
0.1958071825 |
- |
- |
Ataxin-3, putative [Ricinus communis] |
| 19 |
Hb_001329_240 |
0.1985126676 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 20 |
Hb_001959_050 |
0.1998622857 |
- |
- |
PREDICTED: uncharacterized protein LOC103323811 isoform X1 [Prunus mume] |