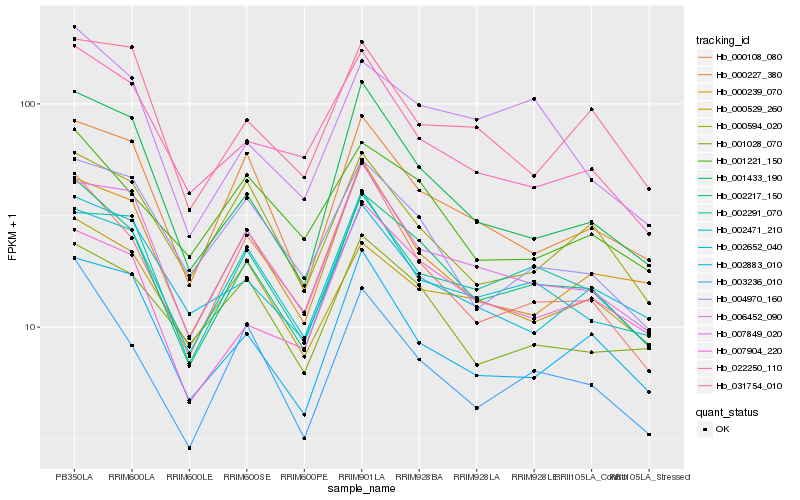
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_150 |
0.0 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 2 |
Hb_003236_010 |
0.0835278405 |
- |
- |
PREDICTED: uncharacterized protein LOC105635957 [Jatropha curcas] |
| 3 |
Hb_007849_020 |
0.0836586137 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 4 |
Hb_002471_210 |
0.0861080663 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 5 |
Hb_001433_190 |
0.0864012023 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 6 |
Hb_002291_070 |
0.0869140228 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Prunus mume] |
| 7 |
Hb_000227_380 |
0.08856959 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 8 |
Hb_002883_010 |
0.0905768342 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006452_090 |
0.0941805354 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5B [Jatropha curcas] |
| 10 |
Hb_000108_080 |
0.0955108528 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 11 |
Hb_000529_260 |
0.0961907273 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 12 |
Hb_007904_220 |
0.0977346749 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_022250_110 |
0.1003356302 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000239_070 |
0.1007980132 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002652_040 |
0.1013585886 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 16 |
Hb_001028_070 |
0.102486981 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 17 |
Hb_031754_010 |
0.1031056338 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_000594_020 |
0.1035760557 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001221_150 |
0.1037045097 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_004970_160 |
0.1039053392 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |