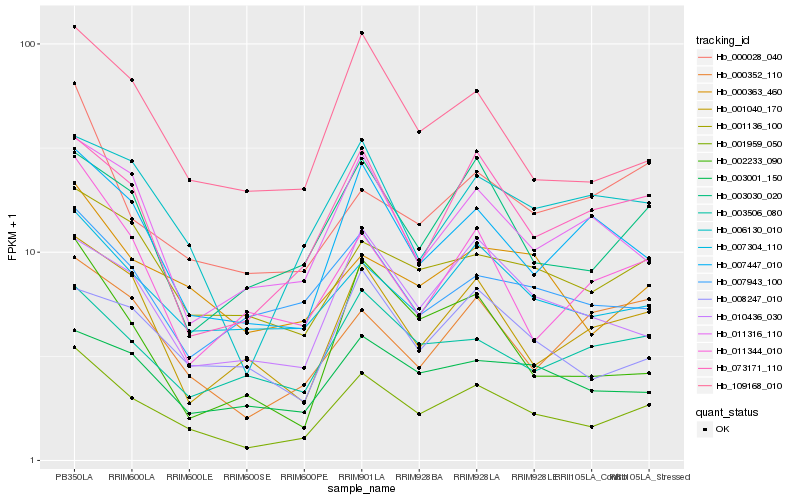
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103323811 isoform X1 [Prunus mume] |
| 2 |
Hb_109168_010 |
0.1174726964 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_007304_110 |
0.1201916305 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 4 |
Hb_007447_010 |
0.1251736726 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003030_020 |
0.1266534665 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |
| 6 |
Hb_073171_110 |
0.1274156319 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 7 |
Hb_001136_100 |
0.1278781184 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 8 |
Hb_000363_460 |
0.1280061054 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_010436_030 |
0.1323364975 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 10 |
Hb_001040_170 |
0.134047121 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_006130_010 |
0.1340986413 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 12 |
Hb_003001_150 |
0.1375360462 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003506_080 |
0.1401212699 |
- |
- |
dipeptidyl peptidase IV, putative [Ricinus communis] |
| 14 |
Hb_011316_110 |
0.1405989236 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 15 |
Hb_008247_010 |
0.1433360016 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_000352_110 |
0.1475999224 |
- |
- |
DNA topoisomerase II, putative [Ricinus communis] |
| 17 |
Hb_007943_100 |
0.1481635037 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_002233_090 |
0.1481968703 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_011344_010 |
0.1489058197 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 20 |
Hb_000028_040 |
0.1496187207 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |