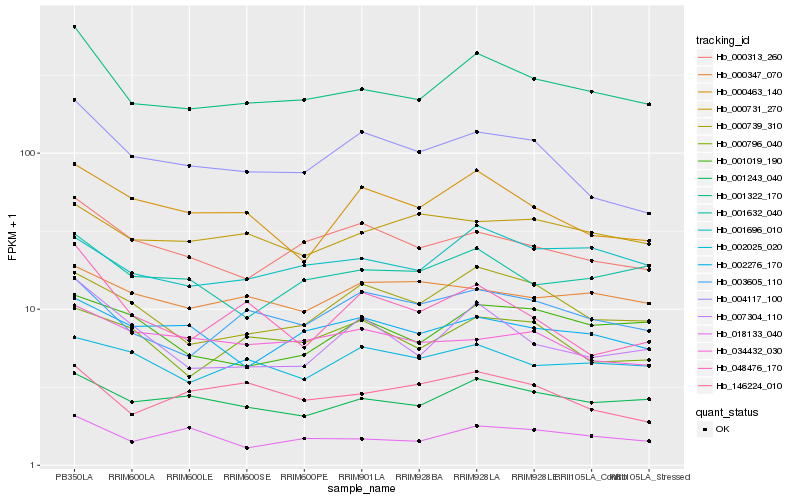
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_170 |
0.0 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 2 |
Hb_001243_040 |
0.088813501 |
- |
- |
PREDICTED: uncharacterized protein LOC104104316 [Nicotiana tomentosiformis] |
| 3 |
Hb_000313_260 |
0.099767773 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 4 |
Hb_018133_040 |
0.099884481 |
- |
- |
PREDICTED: probable apyrase 7 [Jatropha curcas] |
| 5 |
Hb_001696_010 |
0.1009401645 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_001632_040 |
0.1027233888 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_007304_110 |
0.1051783675 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_003605_110 |
0.1054148372 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X1 [Jatropha curcas] |
| 9 |
Hb_004117_100 |
0.1098102754 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000739_310 |
0.1101514272 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002276_170 |
0.1141291286 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000463_140 |
0.1176331392 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001019_190 |
0.1176519086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000731_270 |
0.1194072665 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000796_040 |
0.1199055524 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002025_020 |
0.1204873048 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 17 |
Hb_048476_170 |
0.1219816694 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |
| 18 |
Hb_000347_070 |
0.1234397611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_034432_030 |
0.1236757787 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 20 |
Hb_146224_010 |
0.1240077764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |