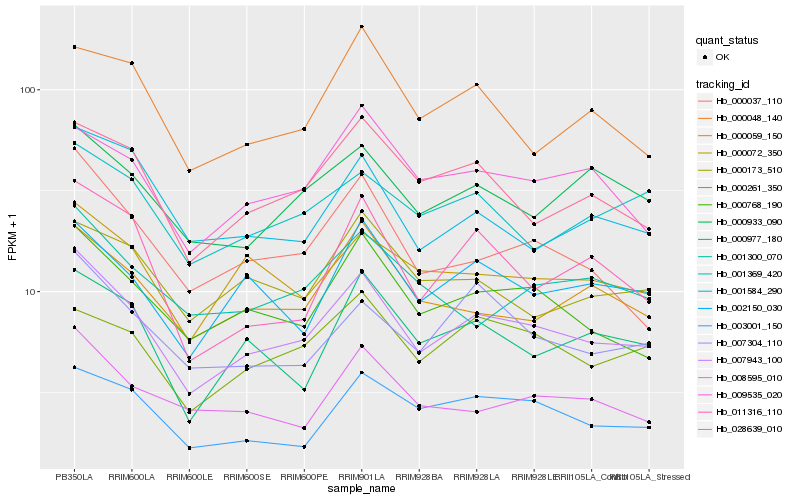
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_100 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_000768_190 |
0.0820977683 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 3 |
Hb_028639_010 |
0.0944226673 |
- |
- |
transporter, putative [Ricinus communis] |
| 4 |
Hb_003001_150 |
0.0977826365 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000037_110 |
0.0979574803 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Populus euphratica] |
| 6 |
Hb_002150_030 |
0.0984328715 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 7 |
Hb_000059_150 |
0.0985633751 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000072_350 |
0.0985642954 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 9 |
Hb_007304_110 |
0.0999574622 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000933_090 |
0.1009183353 |
- |
- |
PREDICTED: probable protein S-acyltransferase 22 [Jatropha curcas] |
| 11 |
Hb_000977_180 |
0.1009370166 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009535_020 |
0.102279397 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 13 |
Hb_008595_010 |
0.1034927905 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000261_350 |
0.1041502173 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011316_110 |
0.1065412484 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 16 |
Hb_001584_290 |
0.1079286609 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 17 |
Hb_001369_420 |
0.1083035633 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 18 |
Hb_000048_140 |
0.1089579493 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 19 |
Hb_001300_070 |
0.1094614012 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 20 |
Hb_000173_510 |
0.1097875478 |
- |
- |
PREDICTED: AP-5 complex subunit mu [Jatropha curcas] |