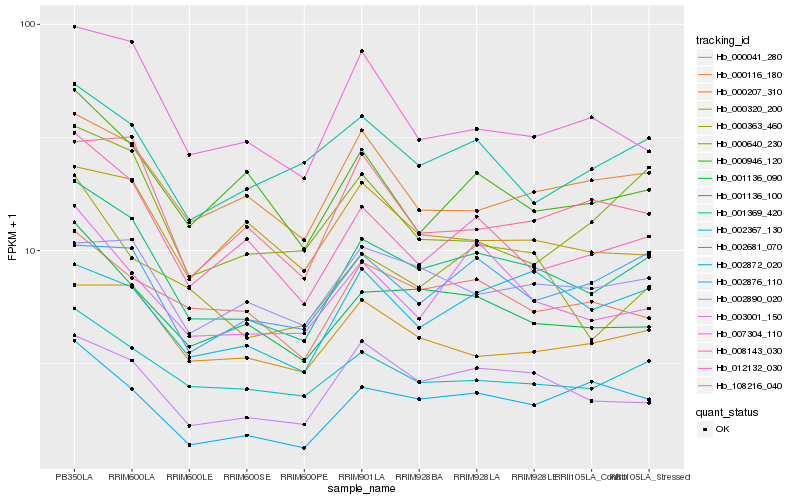
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001136_100 |
0.0 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 2 |
Hb_001136_090 |
0.0878477087 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 3 |
Hb_002367_130 |
0.0887825962 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_012132_030 |
0.0899055016 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 5 |
Hb_003001_150 |
0.0942112565 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_007304_110 |
0.1021805668 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000207_310 |
0.1035884532 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 8 |
Hb_002872_020 |
0.1066864586 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000363_460 |
0.107597881 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000041_280 |
0.1083835969 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 6 [Jatropha curcas] |
| 11 |
Hb_000116_180 |
0.1110092452 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 12 |
Hb_000946_120 |
0.1115004212 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_008143_030 |
0.113235439 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 14 |
Hb_108216_040 |
0.1140523606 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 15 |
Hb_002890_020 |
0.1146917238 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001369_420 |
0.1159476207 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 17 |
Hb_002876_110 |
0.116733615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000640_230 |
0.117115317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002681_070 |
0.1186256364 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000320_200 |
0.1197910503 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |