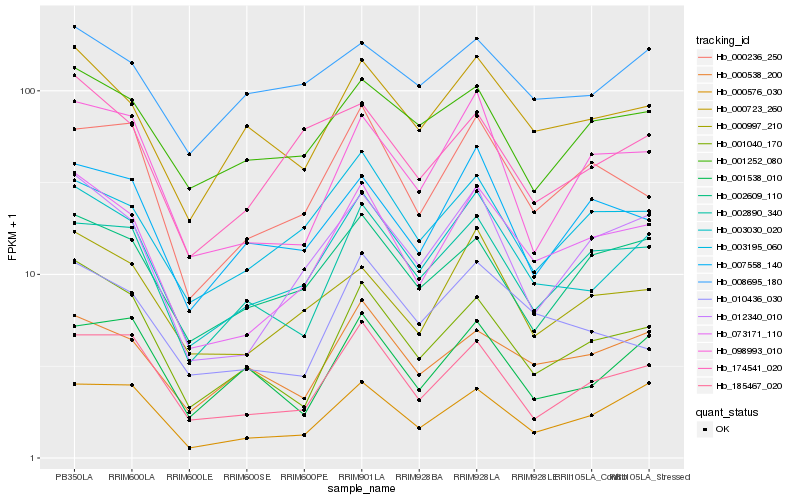
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003030_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |
| 2 |
Hb_073171_110 |
0.086220437 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 3 |
Hb_000723_260 |
0.094761492 |
- |
- |
ethylene insensitive protein, putative [Ricinus communis] |
| 4 |
Hb_185467_020 |
0.1037248159 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_000997_210 |
0.1051477116 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 6 |
Hb_012340_010 |
0.1055970022 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 7 |
Hb_001040_170 |
0.111050655 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_174541_020 |
0.1120549668 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6-like [Populus euphratica] |
| 9 |
Hb_001252_080 |
0.1126825179 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 10 |
Hb_002890_340 |
0.1139047835 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003195_060 |
0.1147102481 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 12 |
Hb_002609_110 |
0.1180027641 |
- |
- |
hypothetical protein JCGZ_08497 [Jatropha curcas] |
| 13 |
Hb_098993_010 |
0.1184515124 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000576_030 |
0.1184520721 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20540 [Jatropha curcas] |
| 15 |
Hb_010436_030 |
0.1203607097 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 16 |
Hb_000236_250 |
0.1209268129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000538_200 |
0.1211650126 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22070-like [Jatropha curcas] |
| 18 |
Hb_007558_140 |
0.1214270533 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 19 |
Hb_001538_010 |
0.1220480746 |
- |
- |
- |
| 20 |
Hb_008695_180 |
0.122114592 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |