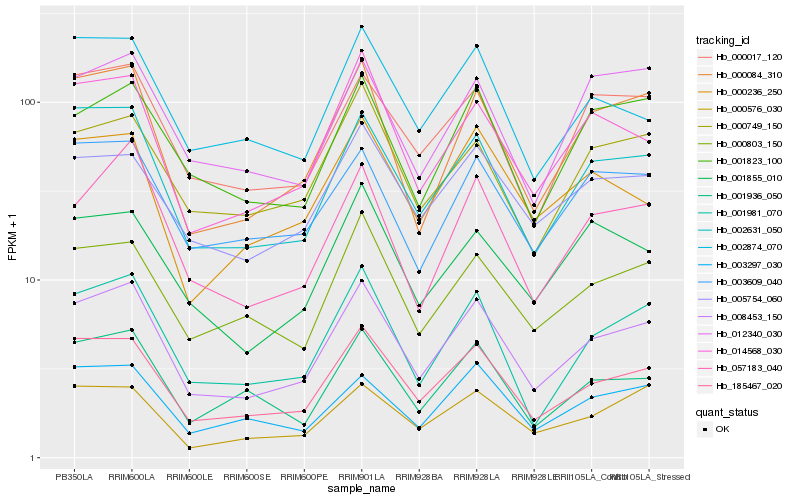
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008453_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X2 [Jatropha curcas] |
| 2 |
Hb_185467_020 |
0.0483193039 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_002631_050 |
0.0618391175 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 4 |
Hb_000084_310 |
0.0714952818 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 5 |
Hb_001981_070 |
0.073731374 |
- |
- |
tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_014568_030 |
0.083791143 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 7 |
Hb_057183_040 |
0.0852412834 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 2 [Jatropha curcas] |
| 8 |
Hb_000803_150 |
0.0867203863 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000017_120 |
0.0871655786 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 10 |
Hb_003297_030 |
0.0918356874 |
- |
- |
PREDICTED: uncharacterized protein LOC105640530 [Jatropha curcas] |
| 11 |
Hb_001936_050 |
0.0939721485 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210-like [Prunus mume] |
| 12 |
Hb_003609_040 |
0.0947960773 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 13 |
Hb_005754_060 |
0.0954159996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001823_100 |
0.0957489994 |
- |
- |
PREDICTED: probable ribosome biogenesis protein RLP24 [Vitis vinifera] |
| 15 |
Hb_000576_030 |
0.0968124577 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20540 [Jatropha curcas] |
| 16 |
Hb_000749_150 |
0.0977420021 |
- |
- |
- |
| 17 |
Hb_002874_070 |
0.0990074561 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 18 |
Hb_001855_010 |
0.1038181093 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_000236_250 |
0.1043544506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012340_030 |
0.1044177499 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |