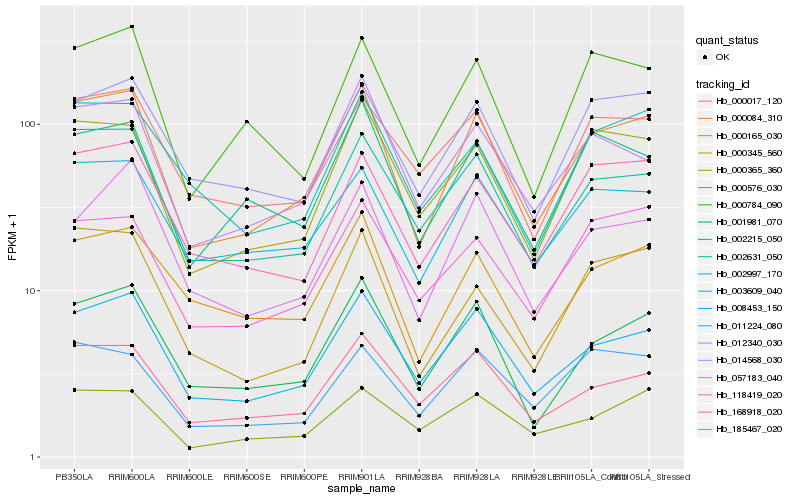
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_310 |
0.0 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 2 |
Hb_008453_150 |
0.0714952818 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002631_050 |
0.0847639835 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 4 |
Hb_001981_070 |
0.0849306682 |
- |
- |
tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000365_360 |
0.0891890419 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_168918_020 |
0.092065017 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 7 |
Hb_185467_020 |
0.0943615529 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 8 |
Hb_003609_040 |
0.0958170539 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 9 |
Hb_014568_030 |
0.098960496 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 10 |
Hb_012340_030 |
0.0991297724 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |
| 11 |
Hb_000165_030 |
0.0991891169 |
- |
- |
PREDICTED: uncharacterized protein LOC105645673 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_560 |
0.0995938643 |
- |
- |
- |
| 13 |
Hb_002215_050 |
0.1001958212 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 14 |
Hb_000576_030 |
0.1004273716 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20540 [Jatropha curcas] |
| 15 |
Hb_118419_020 |
0.1014667722 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Vitis vinifera] |
| 16 |
Hb_000784_090 |
0.1017270071 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_000017_120 |
0.1060403284 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 18 |
Hb_002997_170 |
0.1062400495 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 19 |
Hb_057183_040 |
0.1064188555 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 2 [Jatropha curcas] |
| 20 |
Hb_011224_080 |
0.1064636594 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |