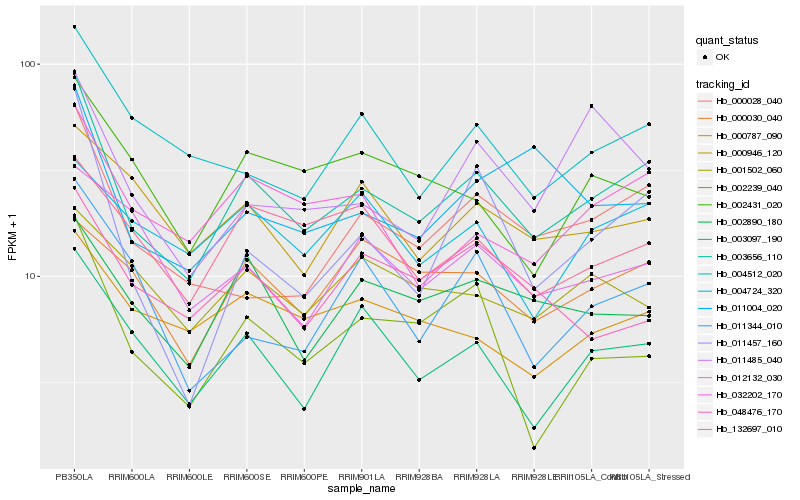
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003656_110 |
0.0 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_032202_170 |
0.127658025 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 3 |
Hb_000028_040 |
0.1284316181 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |
| 4 |
Hb_003097_190 |
0.1350736346 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 5 |
Hb_004512_020 |
0.1390850819 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 6 |
Hb_011457_160 |
0.1412394319 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 7 |
Hb_002890_180 |
0.1421376082 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 8 |
Hb_000787_090 |
0.148811494 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 9 |
Hb_132697_010 |
0.14940925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000946_120 |
0.153663892 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_002431_020 |
0.1537574227 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 12 |
Hb_002239_040 |
0.1539676563 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 13 |
Hb_011004_020 |
0.1547271139 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 14 |
Hb_004724_320 |
0.1567655734 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 15 |
Hb_011344_010 |
0.1575455734 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 16 |
Hb_012132_030 |
0.158925522 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 17 |
Hb_000030_040 |
0.1592860559 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001502_060 |
0.1596198162 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 19 |
Hb_011485_040 |
0.1596816945 |
- |
- |
PREDICTED: probable mitochondrial adenine nucleotide transporter BTL3 [Jatropha curcas] |
| 20 |
Hb_048476_170 |
0.1628279522 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |