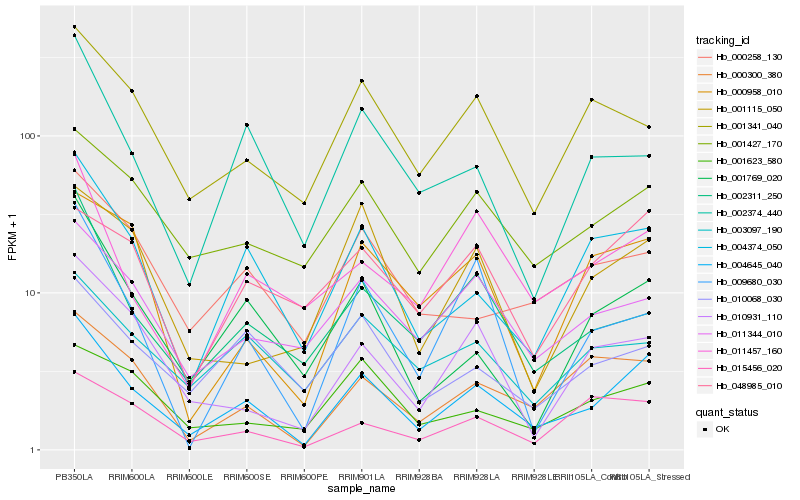
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004645_040 |
0.0 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 2 |
Hb_004374_050 |
0.1541147399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000300_380 |
0.173358454 |
- |
- |
- |
| 4 |
Hb_001769_020 |
0.1735763928 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001427_170 |
0.1746659147 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 6 |
Hb_011344_010 |
0.1836446007 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 7 |
Hb_011457_160 |
0.1856568878 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 8 |
Hb_002311_250 |
0.1874951892 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 9 |
Hb_002374_440 |
0.1916790368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010931_110 |
0.1920742339 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_000258_130 |
0.1937321007 |
- |
- |
- |
| 12 |
Hb_000958_010 |
0.1954580135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010068_030 |
0.1960118768 |
- |
- |
aconitase, putative [Ricinus communis] |
| 14 |
Hb_003097_190 |
0.1962601714 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 15 |
Hb_001341_040 |
0.196405525 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 16 |
Hb_009680_030 |
0.1983736287 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 17 |
Hb_015456_020 |
0.1989225493 |
- |
- |
- |
| 18 |
Hb_048985_010 |
0.1996606907 |
- |
- |
- |
| 19 |
Hb_001623_580 |
0.2037275961 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 20 |
Hb_001115_050 |
0.2052831719 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |