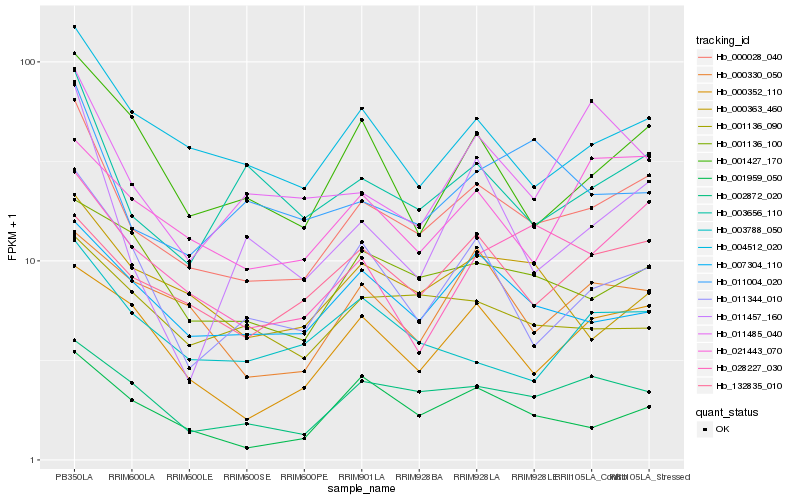
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_040 |
0.0 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |
| 2 |
Hb_004512_020 |
0.1209338922 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 3 |
Hb_003656_110 |
0.1284316181 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_002872_020 |
0.13217173 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_011344_010 |
0.1417340717 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 6 |
Hb_001427_170 |
0.1422984899 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 7 |
Hb_000352_110 |
0.1427557896 |
- |
- |
DNA topoisomerase II, putative [Ricinus communis] |
| 8 |
Hb_132835_010 |
0.1429603772 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_007304_110 |
0.1465736595 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_028227_030 |
0.1476752524 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001959_050 |
0.1496187207 |
- |
- |
PREDICTED: uncharacterized protein LOC103323811 isoform X1 [Prunus mume] |
| 12 |
Hb_001136_100 |
0.151247917 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 13 |
Hb_011004_020 |
0.1519274281 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 14 |
Hb_000363_460 |
0.1544898286 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_000330_050 |
0.1563074378 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 16 |
Hb_021443_070 |
0.156986199 |
- |
- |
PREDICTED: cysteine and histidine-rich domain-containing protein RAR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011457_160 |
0.1595746579 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 18 |
Hb_011485_040 |
0.1612143492 |
- |
- |
PREDICTED: probable mitochondrial adenine nucleotide transporter BTL3 [Jatropha curcas] |
| 19 |
Hb_003788_050 |
0.1613438612 |
- |
- |
PREDICTED: interferon-induced guanylate-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_001136_090 |
0.1617405274 |
- |
- |
Protein PNS1, putative [Ricinus communis] |