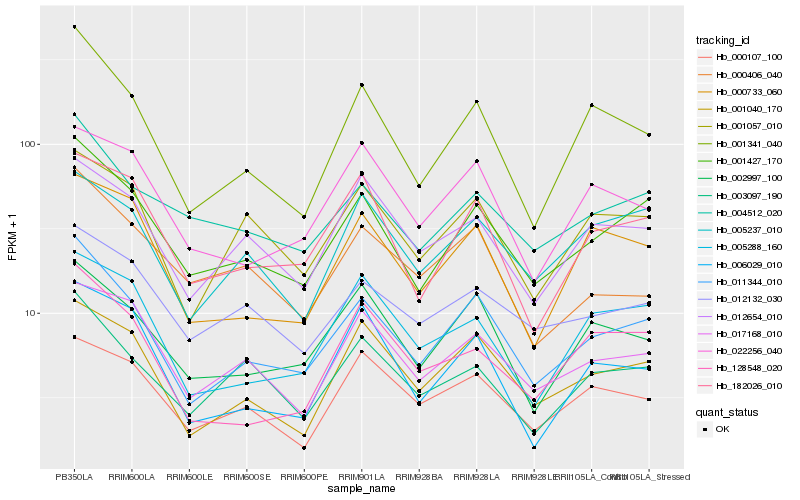
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_010 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 2 |
Hb_001427_170 |
0.0768060344 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 3 |
Hb_001341_040 |
0.0857094334 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 4 |
Hb_001040_170 |
0.0995297493 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_002997_100 |
0.1051563143 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 6 |
Hb_001057_010 |
0.1126089956 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 7 |
Hb_000733_060 |
0.116809769 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_182026_010 |
0.1175626875 |
- |
- |
hypothetical protein CICLE_v10003487mg, partial [Citrus clementina] |
| 9 |
Hb_004512_020 |
0.1175627386 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 10 |
Hb_003097_190 |
0.1180556215 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 11 |
Hb_017168_010 |
0.1210172132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005288_160 |
0.1224369408 |
- |
- |
PREDICTED: uncharacterized protein LOC105647746 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012132_030 |
0.1228572024 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 14 |
Hb_012654_010 |
0.1228902179 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 15 |
Hb_005237_010 |
0.1248367552 |
- |
- |
PREDICTED: uncharacterized protein LOC105630921 [Jatropha curcas] |
| 16 |
Hb_128548_020 |
0.125300991 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 17 |
Hb_006029_010 |
0.1287488181 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 18 |
Hb_022256_040 |
0.1288409535 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase rbrA isoform X2 [Jatropha curcas] |
| 19 |
Hb_000406_040 |
0.1300375916 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 20 |
Hb_000107_100 |
0.1301312058 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |