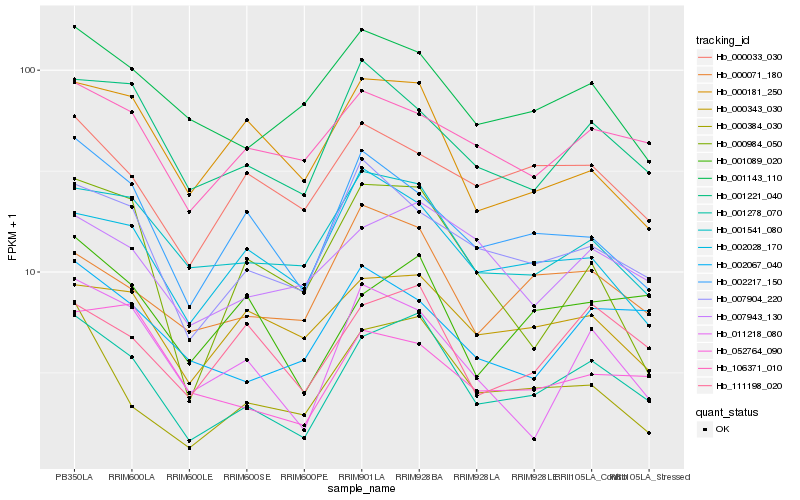
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001278_070 |
0.0 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 2 |
Hb_000384_030 |
0.1439674941 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_111198_020 |
0.14853612 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 4 |
Hb_001089_020 |
0.1524830173 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_007904_220 |
0.1528051344 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_001541_080 |
0.1555443842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002217_150 |
0.1566608048 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 8 |
Hb_007943_130 |
0.1594042967 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 9 |
Hb_001221_040 |
0.1601904032 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 10 |
Hb_011218_080 |
0.160707242 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 11 |
Hb_000071_180 |
0.1664090627 |
- |
- |
PREDICTED: putative DEAD-box ATP-dependent RNA helicase 29 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000343_030 |
0.1667537165 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 13 |
Hb_000984_050 |
0.1701658316 |
- |
- |
sucrose transporter 5 [Hevea brasiliensis] |
| 14 |
Hb_002067_040 |
0.1704033162 |
- |
- |
dihydrodipicolinate reductase, putative [Ricinus communis] |
| 15 |
Hb_001143_110 |
0.1704796262 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 16 |
Hb_002028_170 |
0.1723828442 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 17 |
Hb_052764_090 |
0.1729262744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000181_250 |
0.1754630821 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 19 |
Hb_106371_010 |
0.1755141399 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_000033_030 |
0.1763879318 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |