| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
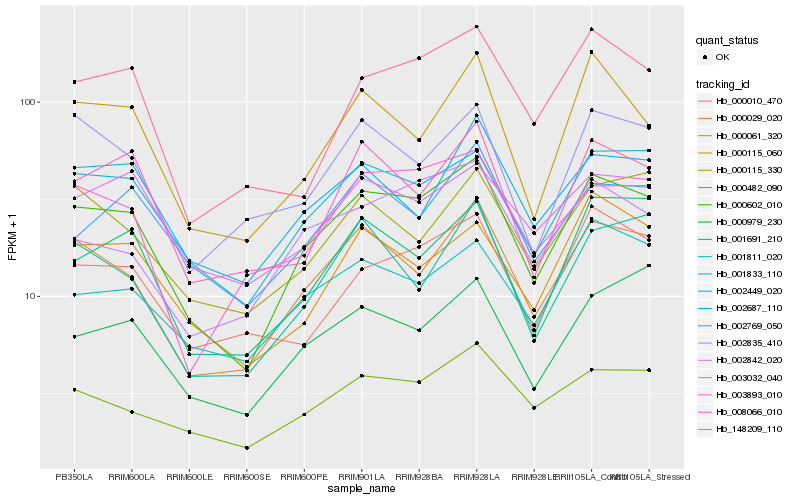
Hb_000602_010 |
0.0 |
- |
- |
PREDICTED: rhomboid-like protease 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000029_020 |
0.0767607963 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 3 |
Hb_002449_020 |
0.0844003345 |
- |
- |
PREDICTED: tRNA (guanine(9)-N1)-methyltransferase [Cucumis sativus] |
| 4 |
Hb_000482_090 |
0.0997220911 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 5 |
Hb_002687_110 |
0.103143974 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C343.04c isoform X1 [Jatropha curcas] |
| 6 |
Hb_001691_210 |
0.1038006513 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 7 |
Hb_000115_060 |
0.1051675323 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 8 |
Hb_002842_020 |
0.1061554281 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 9 |
Hb_003893_010 |
0.1087475494 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 10 |
Hb_000061_320 |
0.1095043076 |
- |
- |
PREDICTED: uncharacterized protein LOC105640575 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001833_110 |
0.109753858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008066_010 |
0.1109180537 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |
| 13 |
Hb_148209_110 |
0.1110451637 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 14 |
Hb_002769_050 |
0.1113430945 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 15 |
Hb_003032_040 |
0.1130215056 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 16 |
Hb_001811_020 |
0.1131437258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000979_230 |
0.1135480079 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |
| 18 |
Hb_000010_470 |
0.1138381371 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_000115_330 |
0.1150026218 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 20 |
Hb_002835_410 |
0.115036155 |
- |
- |
clathrin coat assembly protein ap-1, putative [Ricinus communis] |