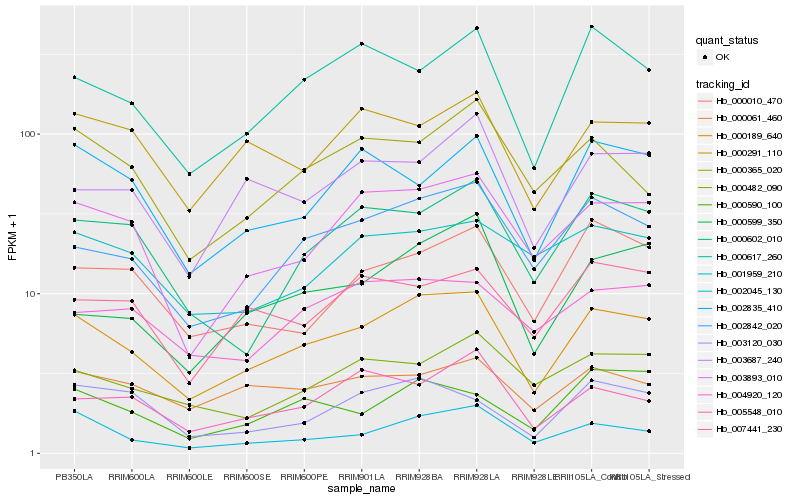
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_640 |
0.0 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 2 |
Hb_002842_020 |
0.1016622229 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 3 |
Hb_003893_010 |
0.1038405568 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 4 |
Hb_002045_130 |
0.1189509034 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 5 |
Hb_000617_260 |
0.1234326601 |
rubber biosynthesis |
Gene Name: SRPP2 |
small rubber particle protein [Hevea brasiliensis] |
| 6 |
Hb_000291_110 |
0.1275277979 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 7 |
Hb_000602_010 |
0.1280046066 |
- |
- |
PREDICTED: rhomboid-like protease 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004920_120 |
0.1280356093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003687_240 |
0.1285984258 |
- |
- |
PREDICTED: cryptochrome-1 [Jatropha curcas] |
| 10 |
Hb_000061_460 |
0.130474904 |
transcription factor |
TF Family: SNF2 |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_007441_230 |
0.1317101813 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_000010_470 |
0.1319272396 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_000482_090 |
0.1325387641 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 14 |
Hb_002835_410 |
0.1326070595 |
- |
- |
clathrin coat assembly protein ap-1, putative [Ricinus communis] |
| 15 |
Hb_000599_350 |
0.1357265823 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_000365_020 |
0.1357315777 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_001959_210 |
0.1362381232 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 18 |
Hb_005548_010 |
0.1379984619 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 19 |
Hb_003120_030 |
0.1380267422 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 20 |
Hb_000590_100 |
0.1380435217 |
- |
- |
- |