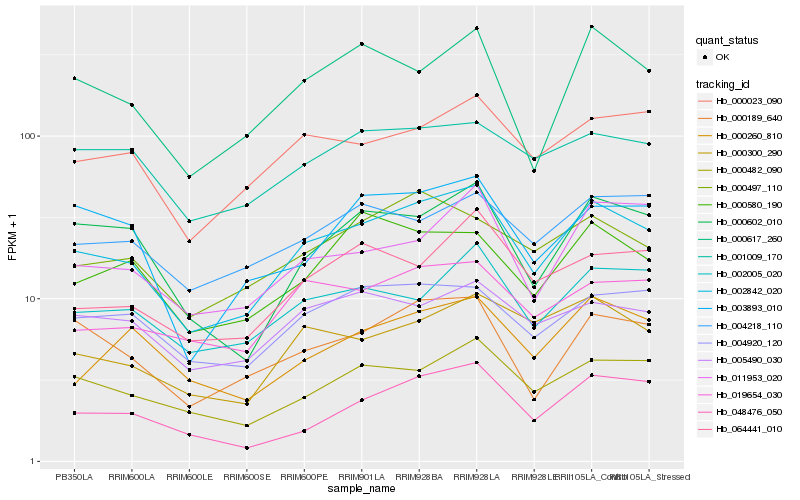
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002842_020 |
0.0 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 2 |
Hb_000482_090 |
0.1011990647 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 3 |
Hb_000189_640 |
0.1016622229 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 4 |
Hb_000023_090 |
0.1044454475 |
- |
- |
mak, putative [Ricinus communis] |
| 5 |
Hb_000602_010 |
0.1061554281 |
- |
- |
PREDICTED: rhomboid-like protease 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_019654_030 |
0.1087341758 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_002005_020 |
0.1090041888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004920_120 |
0.1095017835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000617_260 |
0.1103445566 |
rubber biosynthesis |
Gene Name: SRPP2 |
small rubber particle protein [Hevea brasiliensis] |
| 10 |
Hb_048476_050 |
0.1107945027 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 11 |
Hb_000497_110 |
0.1110261942 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 12 |
Hb_064441_010 |
0.1146056708 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000300_290 |
0.1151772834 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 14 |
Hb_001009_170 |
0.1171579465 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_003893_010 |
0.1175318928 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 16 |
Hb_005490_030 |
0.1179645838 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X3 [Jatropha curcas] |
| 17 |
Hb_011953_020 |
0.118798046 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000260_810 |
0.119387533 |
- |
- |
- |
| 19 |
Hb_000580_190 |
0.1194937467 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 20 |
Hb_004218_110 |
0.1195370802 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |