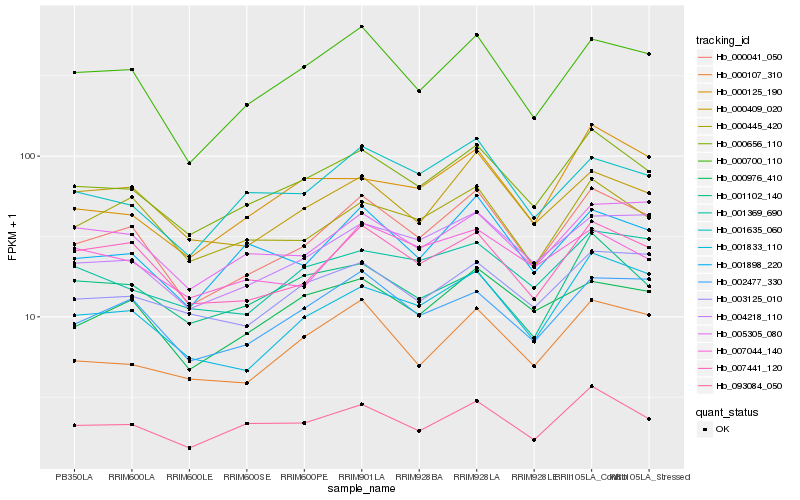
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_110 |
0.0 |
- |
- |
nucleolysin tia-1, putative [Ricinus communis] |
| 2 |
Hb_093084_050 |
0.0448014726 |
- |
- |
PREDICTED: uncharacterized protein LOC105640739 [Jatropha curcas] |
| 3 |
Hb_000041_050 |
0.0626741533 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 4 |
Hb_001102_140 |
0.073965275 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 2-like [Jatropha curcas] |
| 5 |
Hb_000700_110 |
0.0808143291 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 6 |
Hb_003125_010 |
0.0812415726 |
- |
- |
PREDICTED: uncharacterized protein LOC105640205 [Jatropha curcas] |
| 7 |
Hb_001369_690 |
0.0812788602 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 8 |
Hb_001898_220 |
0.08309861 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004218_110 |
0.0834224754 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 10 |
Hb_001635_060 |
0.0834448579 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 11 |
Hb_000976_410 |
0.084872845 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 12 |
Hb_000125_190 |
0.0857459687 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007441_120 |
0.0858933189 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000445_420 |
0.0863034133 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_000107_310 |
0.087538703 |
- |
- |
PREDICTED: pseudouridine-metabolizing bifunctional protein C1861.05 [Jatropha curcas] |
| 16 |
Hb_002477_330 |
0.0902455063 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_001833_110 |
0.0903016058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005305_080 |
0.0908544786 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000409_020 |
0.0920363565 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 20 |
Hb_007044_140 |
0.0927478337 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta [Jatropha curcas] |