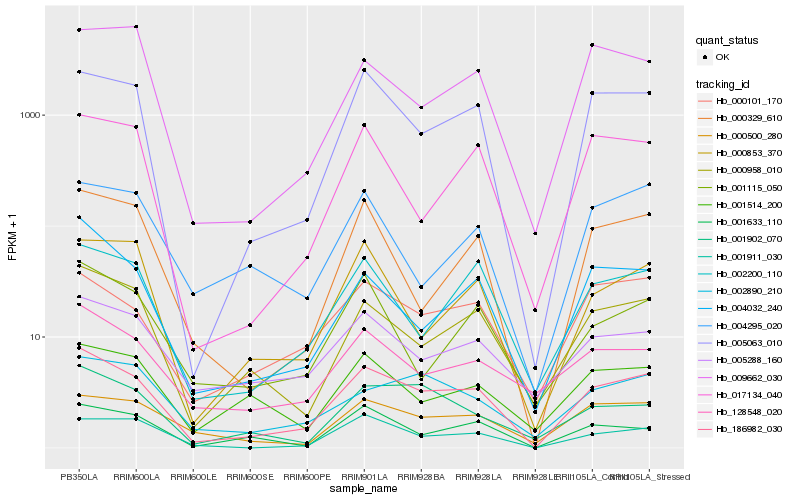
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186982_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005063_010 |
0.117403637 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 3 |
Hb_000958_010 |
0.1359654538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000101_170 |
0.1418966342 |
- |
- |
- |
| 5 |
Hb_002200_110 |
0.1506641017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004032_240 |
0.151272825 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 7 |
Hb_001911_030 |
0.1518235233 |
- |
- |
PREDICTED: disease resistance RPP8-like protein 3 [Jatropha curcas] |
| 8 |
Hb_000329_610 |
0.1582492407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000500_280 |
0.1583742821 |
- |
- |
Brassinosteroid-regulated protein BRU1 precursor, putative [Ricinus communis] |
| 10 |
Hb_128548_020 |
0.1601316549 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 11 |
Hb_005288_160 |
0.1608688911 |
- |
- |
PREDICTED: uncharacterized protein LOC105647746 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001633_110 |
0.1617435646 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000853_370 |
0.1640076543 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 14 |
Hb_017134_040 |
0.1649077752 |
- |
- |
hypothetical protein AALP_AA3G042000 [Arabis alpina] |
| 15 |
Hb_001902_070 |
0.1649365742 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_001115_050 |
0.1691043757 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 17 |
Hb_002890_210 |
0.1699656586 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 18 |
Hb_001514_200 |
0.1716473168 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 19 |
Hb_009662_030 |
0.1780642457 |
- |
- |
hypothetical protein JCGZ_15748 [Jatropha curcas] |
| 20 |
Hb_004295_020 |
0.1798358907 |
- |
- |
thioredoxin II-type 1 [Hevea brasiliensis] |