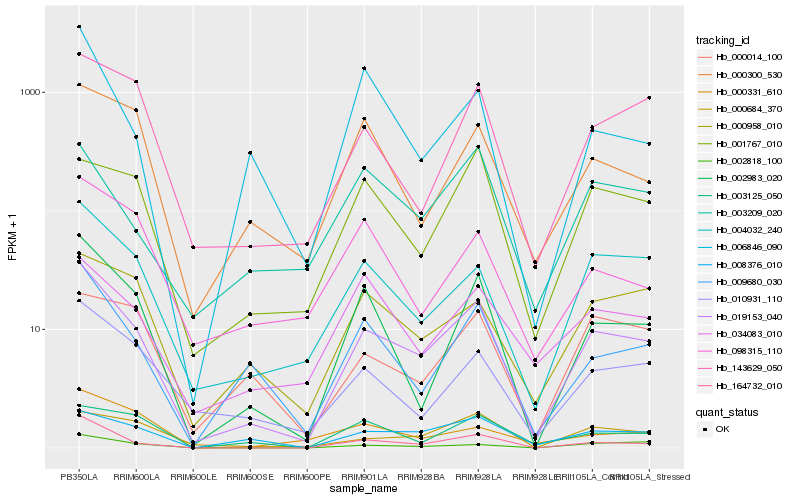
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019153_040 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_164732_010 |
0.142443131 |
- |
- |
- |
| 3 |
Hb_002983_020 |
0.1486216124 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Populus euphratica] |
| 4 |
Hb_004032_240 |
0.1507921768 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_009680_030 |
0.1588517453 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 6 |
Hb_010931_110 |
0.1683410611 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_008376_010 |
0.1756449127 |
- |
- |
- |
| 8 |
Hb_143629_050 |
0.1808891661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002818_100 |
0.1836403387 |
- |
- |
hypothetical protein JCGZ_14122 [Jatropha curcas] |
| 10 |
Hb_000684_370 |
0.1881909651 |
- |
- |
- |
| 11 |
Hb_000958_010 |
0.1901296263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003125_050 |
0.1931102005 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000300_530 |
0.1943680649 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0286299-like [Gossypium raimondii] |
| 14 |
Hb_034083_010 |
0.1983256225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001767_010 |
0.1983989999 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 16 |
Hb_006846_090 |
0.1985167039 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 17 |
Hb_003209_020 |
0.2039345763 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 18 |
Hb_098315_110 |
0.2093967413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000331_610 |
0.2099752621 |
- |
- |
hypothetical protein B456_003G108600 [Gossypium raimondii] |
| 20 |
Hb_000014_100 |
0.2103619265 |
- |
- |
- |